CCR2+ monocytes replenish border-associated macrophages in the diseased mouse brain
- PMID: 38625796
- PMCID: PMC11105166
- DOI: 10.1016/j.celrep.2024.114120
CCR2+ monocytes replenish border-associated macrophages in the diseased mouse brain
Abstract
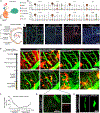
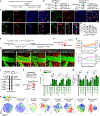
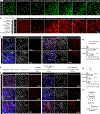
Border-associated macrophages (BAMs) are tissue-resident macrophages that reside at the border of the central nervous system (CNS). Since BAMs originate from yolk sac progenitors that do not persist after birth, the means by which this population of cells is maintained is not well understood. Using two-photon microscopy and multiple lineage-tracing strategies, we determine that CCR2+ monocytes are significant contributors to BAM populations following disruptions of CNS homeostasis in adult mice. After BAM depletion, while the residual BAMs possess partial self-repopulation capability, the CCR2+ monocytes are a critical source of the repopulated BAMs. In addition, we demonstrate the existence of CCR2+ monocyte-derived long-lived BAMs in a brain compression model and in a sepsis model after the initial disruption of homeostasis. Our study reveals that the short-lived CCR2+ monocytes transform into long-lived BAM-like cells at the CNS border and subsequently contribute to BAM populations.
Keywords: BAM; CCR2; CP: Immunology; CP: Neuroscience; LPS; border-associated macrophages; brain compression; csf1; meningitis; microglia; monocyte; repopulation.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

