Exposing the molecular heterogeneity of glycosylated biotherapeutics
- PMID: 38627419
- PMCID: PMC11021452
- DOI: 10.1038/s41467-024-47693-8
Exposing the molecular heterogeneity of glycosylated biotherapeutics
Abstract
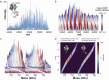
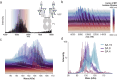
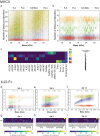
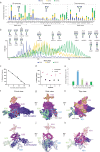
The heterogeneity inherent in today's biotherapeutics, especially as a result of heavy glycosylation, can affect a molecule's safety and efficacy. Characterizing this heterogeneity is crucial for drug development and quality assessment, but existing methods are limited in their ability to analyze intact glycoproteins or other heterogeneous biotherapeutics. Here, we present an approach to the molecular assessment of biotherapeutics that uses proton-transfer charge-reduction with gas-phase fractionation to analyze intact heterogeneous and/or glycosylated proteins by mass spectrometry. The method provides a detailed landscape of the intact molecular weights present in biotherapeutic protein preparations in a single experiment. For glycoproteins in particular, the method may offer insights into glycan composition when coupled with a suitable bioinformatic strategy. We tested the approach on various biotherapeutic molecules, including Fc-fusion, VHH-fusion, and peptide-bound MHC class II complexes to demonstrate efficacy in measuring the proteoform-level diversity of biotherapeutics. Notably, we inferred the glycoform distribution for hundreds of molecular weights for the eight-times glycosylated fusion drug IL22-Fc, enabling correlations between glycoform sub-populations and the drug's pharmacological properties. Our method is broadly applicable and provides a powerful tool to assess the molecular heterogeneity of emerging biotherapeutics.
© 2024. The Author(s).
Conflict of interest statement
L.S., W.P., M.I.B., T.B., P.D., C.T., S.S., T.B., D.D., J.G., A.E., N.A., M.M. and W.S. are employed by Genentech, Inc., a for-profit company that produces and markets therapeutics. C.M., J.H., J.E.P.S., R.M. and R.H. are employed by Thermo Fisher Scientific, a for-profit company that manufactures and sells mass spectrometry equipment. The remaining authors declare no competing interests.
Figures

References
-
- Rathore, A. S. & Winkle, H. Quality by design for biopharmaceuticals. Nat. Biotechnol.27, 26–34 (2009). - PubMed
-
- Walsh, G. Biopharmaceutical benchmarks 2018. Nat. Biotechnol.36, 1136–1145 (2018). - PubMed
-
- Zhang, P. et al. Challenges of glycosylation analysis and control: an integrated approach to producing optimal and consistent therapeutic drugs. Drug Discov. Today21, 740–765 (2016). - PubMed
-
- Silsirivanit, A. Chapter Five Glycosylation markers in cancer. Adv. Clin. Chem.89, 189–213 (2019). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

