Molecular basis of Gabija anti-phage supramolecular assemblies
- PMID: 38627580
- PMCID: PMC11418746
- DOI: 10.1038/s41594-024-01283-w
Molecular basis of Gabija anti-phage supramolecular assemblies
Abstract
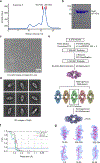
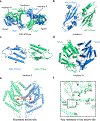
As one of the most prevalent anti-phage defense systems in prokaryotes, Gabija consists of a Gabija protein A (GajA) and a Gabija protein B (GajB). The assembly and function of the Gabija system remain unclear. Here we present cryo-EM structures of Bacillus cereus GajA and GajAB complex, revealing tetrameric and octameric assemblies, respectively. In the center of the complex, GajA assembles into a tetramer, which recruits two sets of GajB dimer at opposite sides of the complex, resulting in a 4:4 GajAB supramolecular complex for anti-phage defense. Further biochemical analysis showed that GajA alone is sufficient to cut double-stranded DNA and plasmid DNA, which can be inhibited by ATP. Unexpectedly, the GajAB displays enhanced activity for plasmid DNA, suggesting a role of substrate selection by GajB. Together, our study defines a framework for understanding anti-phage immune defense by the GajAB complex.
© 2024. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing interests
All authors declare they have no competing interests.
Figures











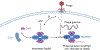
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

