Identification of N7-methylguanosine-related miRNAs as potential biomarkers for prognosis and drug response in breast cancer
- PMID: 38628712
- PMCID: PMC11017060
- DOI: 10.1016/j.heliyon.2024.e29326
Identification of N7-methylguanosine-related miRNAs as potential biomarkers for prognosis and drug response in breast cancer
Abstract
Objectives: The impact of N7-methylguanosine (m7G) on tumor progression and the regulatory role of microRNAs (miRNAs) in immune function significantly influence breast cancer (BC) prognosis. Investigating the interplay between m7G modification and miRNAs provides novel insights for assessing prognostics and drug responses in BC.
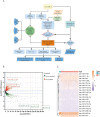
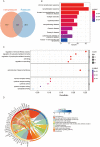
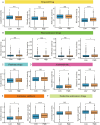
Materials and methods: RNA sequences (miRNA and mRNA profiles) and clinical data for BC were acquired from the Cancer Genome Atlas (TCGA) database. A miRNA signature associated with 15 m7G in this cohort was identified using Cox regression and LASSO. The risk score model was evaluated using Kaplan-Meier and time-dependent ROC analysis, categorizing patients into high-risk and low-risk groups. Functional enrichment analyses were conducted to explore potential pathways. The immune system, including scores, cell infiltration, function, and drug sensitivity, was examined and compared between high-risk and low-risk groups. A nomogram that combines risk scores and clinical factors was developed and validated. Single-sample gene set enrichment analysis (ssGSEA) was employed to explore m7G-related miRNA signatures and immune cell relationships in the tumor microenvironment. Additionally, drug susceptibility was compared between risk groups.
Results: Fifteen m7G-related miRNAs were independently correlated with overall survival (OS) in BC patients. Time-dependent ROC analysis yielded area under the curve (AUC) values of 0.742, 0.726, and 0.712 for predicting 3-, 5-, and 10-year survival rates, respectively. The Kaplan-Meier analysis revealed a significant disparity in OS between the high-risk and low-risk groups (p = 1.3e-6). Multiple regression identified the risk score as a significant independent prognostic factor. An excellent calibration nomogram with a C-index of 0.785 (95 % CI: 0.728-0.843) was constructed. In immune analysis, low-risk patients exhibited heightened immune function and increased responsiveness to immunotherapy and chemotherapy compared to high-risk patients.
Conclusion: This study systematically analyzed m7G-related miRNAs and revealed their regulatory mechanisms concerning the tumor microenvironment (TME), pathology, and the prognosis of BC patient. Based on these miRNAs, a prognostic model and nomogram were developed for BC patients, facilitating prognostic assessments. These findings can also assist in predicting treatment responses and guiding medication selection.
Keywords: Breast cancer; Drug response; N7-methylguanosine; Prognosis; microRNA.
© 2024 Published by Elsevier Ltd.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures




Similar articles
-
Construction of prognostic signature of breast cancer based on N7-Methylguanosine-Related LncRNAs and prediction of immune response.Front Genet. 2022 Oct 24;13:991162. doi: 10.3389/fgene.2022.991162. eCollection 2022. Front Genet. 2022. PMID: 36353118 Free PMC article.
-
Establishment and assessment of an oral squamous cell carcinoma N7-methylguanosine methyltransferase associated microRNA prognostic model.J Cancer. 2024 Sep 30;15(18):6022-6037. doi: 10.7150/jca.98350. eCollection 2024. J Cancer. 2024. PMID: 39440068 Free PMC article.
-
A novel m7G-related miRNA prognostic signature for predicting clinical outcome and immune microenvironment in colon cancer.J Cancer. 2024 Oct 7;15(18):6086-6102. doi: 10.7150/jca.99173. eCollection 2024. J Cancer. 2024. PMID: 39440054 Free PMC article.
-
Construction and validation of a prognostic signature for mucinous colonic adenocarcinoma based on N7-methylguanosine-related long non-coding RNAs.J Gastrointest Oncol. 2024 Feb 29;15(1):203-219. doi: 10.21037/jgo-23-980. Epub 2024 Feb 28. J Gastrointest Oncol. 2024. PMID: 38482248 Free PMC article.
-
N7-methylguanosine modification in cancers: from mechanisms to therapeutic potential.J Hematol Oncol. 2025 Jan 29;18(1):12. doi: 10.1186/s13045-025-01665-7. J Hematol Oncol. 2025. PMID: 39881381 Free PMC article. Review.
References
-
- Peng D., Kryczek I., Nagarsheth N., Zhao L., Wei S., Wang W., Sun Y., Zhao E., Vatan L., Szeliga W., Kotarski J., Tarkowski R., Dou Y., Cho K., Hensley-Alford S., Munkarah A., Liu R., Zou W. Epigenetic silencing of TH1-type chemokines shapes tumour immunity and immunotherapy. Nature. 2015;527:249–253. doi: 10.1038/nature15520. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

