Lethal phenotypes in Mendelian disorders
- PMID: 38629401
- PMCID: PMC11232373
- DOI: 10.1016/j.gim.2024.101141
Lethal phenotypes in Mendelian disorders
Abstract
Purpose: Existing resources that characterize the essentiality status of genes are based on either proliferation assessment in human cell lines, viability evaluation in mouse knockouts, or constraint metrics derived from human population sequencing studies. Several repositories document phenotypic annotations for rare disorders; however, there is a lack of comprehensive reporting on lethal phenotypes.
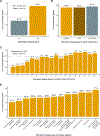
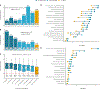
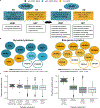
Methods: We queried Online Mendelian Inheritance in Man for terms related to lethality and classified all Mendelian genes according to the earliest age of death recorded for the associated disorders, from prenatal death to no reports of premature death. We characterized the genes across these lethality categories, examined the evidence on viability from mouse models and explored how this information could be used for novel gene discovery.
Results: We developed the Lethal Phenotypes Portal to showcase this curated catalog of human essential genes. Differences in the mode of inheritance, physiological systems affected, and disease class were found for genes in different lethality categories, as well as discrepancies between the lethal phenotypes observed in mouse and human.
Conclusion: We anticipate that this resource will aid clinicians in the diagnosis of early lethal conditions and assist researchers in investigating the properties that make these genes essential for human development.
Keywords: Essential genes; Lethal mouse knockouts; Lethal phenotypes; Mendelian disorders; Novel gene discovery.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of Interest The authors declare no conflicts of interest.
Figures



Update of
-
Lethal phenotypes in Mendelian disorders.medRxiv [Preprint]. 2024 Jan 13:2024.01.12.24301168. doi: 10.1101/2024.01.12.24301168. medRxiv. 2024. Update in: Genet Med. 2024 Jul;26(7):101141. doi: 10.1016/j.gim.2024.101141. PMID: 38260283 Free PMC article. Updated. Preprint.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

