A 3D atlas of the human developing pancreas to explore progenitor proliferation and differentiation
- PMID: 38630142
- PMCID: PMC11058870
- DOI: 10.1007/s00125-024-06143-2
A 3D atlas of the human developing pancreas to explore progenitor proliferation and differentiation
Abstract
Aims/hypothesis: Rodent pancreas development has been described in great detail. On the other hand, there are still gaps in our understanding of the developmental trajectories of pancreatic cells during human ontogenesis. Here, our aim was to map the spatial and chronological dynamics of human pancreatic cell differentiation and proliferation by using 3D imaging of cleared human embryonic and fetal pancreases.
Methods: We combined tissue clearing with light-sheet fluorescence imaging in human embryonic and fetal pancreases during the first trimester of pregnancy. In addition, we validated an explant culture system enabling in vitro proliferation of pancreatic progenitors to determine the mitogenic effect of candidate molecules.
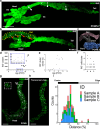
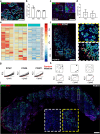
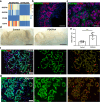
Results: We detected the first insulin-positive cells as early as five post-conceptional weeks, two weeks earlier than previously observed. We observed few insulin-positive clusters at five post-conceptional weeks (mean ± SD 9.25±5.65) with a sharp increase to 11 post-conceptional weeks (4307±152.34). We identified a central niche as the location of onset of the earliest insulin cell production and detected extra-pancreatic loci within the adjacent developing gut. Conversely, proliferating pancreatic progenitors were located in the periphery of the epithelium, suggesting the existence of two separated pancreatic niches for differentiation and proliferation. Additionally, we observed that the proliferation ratio of progenitors ranged between 20% and 30%, while for insulin-positive cells it was 1%. We next unveiled a mitogenic effect of the platelet-derived growth factor AA isoform (PDGFAA) in progenitors acting through the pancreatic mesenchyme by increasing threefold the number of proliferating progenitors.
Conclusions/interpretation: This work presents a first 3D atlas of the human developing pancreas, charting both endocrine and proliferating cells across early development.
Keywords: Human fetal pancreas; Insulin-producing cells; Light-sheet fluorescence microscopy; PDGF signalling; Pancreatic progenitors; Proliferating cells; Type 1 diabetes.
© 2024. The Author(s).
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

