A methodology for specific disruption of microtubule polymerization into dendritic spines
- PMID: 38630519
- PMCID: PMC11238079
- DOI: 10.1091/mbc.E24-02-0093
A methodology for specific disruption of microtubule polymerization into dendritic spines
Abstract
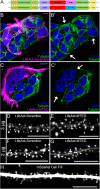
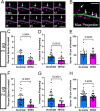
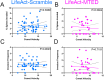
Dendritic spines, the mushroom-shaped extensions along dendritic shafts of excitatory neurons, are critical for synaptic function and are one of the first neuronal structures disrupted in neurodevelopmental and neurodegenerative diseases. Microtubule (MT) polymerization into dendritic spines is an activity-dependent process capable of affecting spine shape and function. Studies have shown that MT polymerization into spines occurs specifically in spines undergoing plastic changes. However, discerning the function of MT invasion of dendritic spines requires the specific inhibition of MT polymerization into spines, while leaving MT dynamics in the dendritic shaft, synaptically connected axons and associated glial cells intact. This is not possible with the unrestricted, bath application of pharmacological compounds. To specifically disrupt MT entry into spines we coupled a MT elimination domain (MTED) from the Efa6 protein to the actin filament-binding peptide LifeAct. LifeAct was chosen because actin filaments are highly concentrated in spines and are necessary for MT invasions. Temporally controlled expression of this LifeAct-MTED construct inhibits MT entry into dendritic spines, while preserving typical MT dynamics in the dendrite shaft. Expression of this construct will allow for the determination of the function of MT invasion of spines and more broadly, to discern how MT-actin interactions affect cellular processes.
Figures



Update of
-
A Methodology for Specific Disruption of Microtubules in Dendritic Spines.bioRxiv [Preprint]. 2024 Mar 4:2024.03.04.583370. doi: 10.1101/2024.03.04.583370. bioRxiv. 2024. Update in: Mol Biol Cell. 2024 Jun 1;35(6):mr3. doi: 10.1091/mbc.E24-02-0093. PMID: 38496454 Free PMC article. Updated. Preprint.
Similar articles
-
A Methodology for Specific Disruption of Microtubules in Dendritic Spines.bioRxiv [Preprint]. 2024 Mar 4:2024.03.04.583370. doi: 10.1101/2024.03.04.583370. bioRxiv. 2024. Update in: Mol Biol Cell. 2024 Jun 1;35(6):mr3. doi: 10.1091/mbc.E24-02-0093. PMID: 38496454 Free PMC article. Updated. Preprint.
-
Synaptic regulation of microtubule dynamics in dendritic spines by calcium, F-actin, and drebrin.J Neurosci. 2013 Oct 16;33(42):16471-82. doi: 10.1523/JNEUROSCI.0661-13.2013. J Neurosci. 2013. PMID: 24133252 Free PMC article.
-
BDNF-induced increase of PSD-95 in dendritic spines requires dynamic microtubule invasions.J Neurosci. 2011 Oct 26;31(43):15597-603. doi: 10.1523/JNEUROSCI.2445-11.2011. J Neurosci. 2011. PMID: 22031905 Free PMC article.
-
Dendritic spine actin dynamics in neuronal maturation and synaptic plasticity.Cytoskeleton (Hoboken). 2016 Sep;73(9):435-41. doi: 10.1002/cm.21280. Epub 2016 Mar 4. Cytoskeleton (Hoboken). 2016. PMID: 26849484 Review.
-
Actin filaments and microtubules in dendritic spines.J Neurochem. 2013 Jul;126(2):155-64. doi: 10.1111/jnc.12313. Epub 2013 Jun 11. J Neurochem. 2013. PMID: 23692384 Review.
Cited by
-
Cerebral Hypoxia-Induced Molecular Alterations and Their Impact on the Physiology of Neurons and Dendritic Spines: A Comprehensive Review.Cell Mol Neurobiol. 2024 Aug 6;44(1):58. doi: 10.1007/s10571-024-01491-4. Cell Mol Neurobiol. 2024. PMID: 39105862 Free PMC article. Review.
-
Emerging roles for tubulin PTMs in neuronal function and neurodegenerative disease.Curr Opin Neurobiol. 2025 Feb;90:102971. doi: 10.1016/j.conb.2025.102971. Epub 2025 Jan 24. Curr Opin Neurobiol. 2025. PMID: 39862522 Review.
References
-
- Bindels DS, Haarbosch L, van Weeren L, Postma M, Wiese KE, Mastop M, Aumonier S, Gotthard G, Royant A, Hink MA, Gadella TW, Jr. (2017). mScarlet: a bright monomeric red fluorescent protein for cellular imaging. Nat Methods 14, 53–56. - PubMed
-
- Borowiak M, Nahaboo W, Reynders M, Nekolla K, Jalinot P, Hasserodt J, Rehberg M, Delattre M, Zahler S, Vollmar A, et al. (2015). Photoswitchable Inhibitors of Microtubule Dynamics Optically Control Mitosis and Cell Death. Cell 162, 403–411. - PubMed
-
- Dent EW, Meiri KF (1992). GAP-43 phosphorylation is dynamically regulated in individual growth cones. J Neurobiol 23, 1037–1053. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

