A macroevolutionary role for chromosomal fusion and fission in Erebia butterflies
- PMID: 38630820
- PMCID: PMC11023530
- DOI: 10.1126/sciadv.adl0989
A macroevolutionary role for chromosomal fusion and fission in Erebia butterflies
Abstract
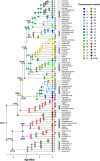
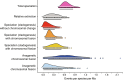
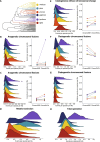
The impact of large-scale chromosomal rearrangements, such as fusions and fissions, on speciation is a long-standing conundrum. We assessed whether bursts of change in chromosome numbers resulting from chromosomal fusion or fission are related to increased speciation rates in Erebia, one of the most species-rich and karyotypically variable butterfly groups. We established a genome-based phylogeny and used state-dependent birth-death models to infer trajectories of karyotype evolution. We demonstrated that rates of anagenetic chromosomal changes (i.e., along phylogenetic branches) exceed cladogenetic changes (i.e., at speciation events), but, when cladogenetic changes occur, they are mostly associated with chromosomal fissions rather than fusions. We found that the relative importance of fusion and fission differs among Erebia clades of different ages and that especially in younger, more karyotypically diverse clades, speciation is more frequently associated with cladogenetic chromosomal changes. Overall, our results imply that chromosomal fusions and fissions have contrasting macroevolutionary roles and that large-scale chromosomal rearrangements are associated with bursts of species diversification.
Figures

References
-
- J. A. Coyne, H. A. Orr, Speciation. (Sinauer Associates, 2004).
-
- White M. J. D., Models of speciation. Science 159, 1065–1070 (1968). - PubMed
-
- M. King, Species Evolution: The Role of Chromosome Change (Cambridge Univ. Press, 1995).
-
- Faria R., Navarro A., Chromosomal speciation revisited: Rearranging theory with pieces of evidence. Trends Ecol. Evol. 25, 660–669 (2010). - PubMed

