Predominance of multidrug-resistant Salmonella Typhi genotype 4.3.1 with low-level ciprofloxacin resistance in Zanzibar
- PMID: 38630840
- PMCID: PMC11057722
- DOI: 10.1371/journal.pntd.0012132
Predominance of multidrug-resistant Salmonella Typhi genotype 4.3.1 with low-level ciprofloxacin resistance in Zanzibar
Abstract
Background: Typhoid fever is a common cause of febrile illness in low- and middle-income countries. While multidrug-resistant (MDR) Salmonella Typhi (S. Typhi) has spread globally, fluoroquinolone resistance has mainly affected Asia.
Methods: Consecutively, 1038 blood cultures were obtained from patients of all age groups with fever and/or suspicion of serious systemic infection admitted at Mnazi Mmoja Hospital, Zanzibar in 2015-2016. S. Typhi were analyzed with antimicrobial susceptibility testing and with short read (61 strains) and long read (9 strains) whole genome sequencing, including three S. Typhi strains isolated in a pilot study 2012-2013.
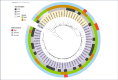
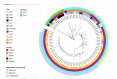
Results: Sixty-three S. Typhi isolates (98%) were MDR carrying blaTEM-1B, sul1 and sul2, dfrA7 and catA1 genes. Low-level ciprofloxacin resistance was detected in 69% (43/62), with a single gyrase mutation gyrA-D87G in 41 strains, and a single gyrA-S83F mutation in the non-MDR strain. All isolates were susceptible to ceftriaxone and azithromycin. All MDR isolates belonged to genotype 4.3.1 lineage I (4.3.1.1), with the antimicrobial resistance determinants located on a composite transposon integrated into the chromosome. Phylogenetically, the MDR subgroup with ciprofloxacin resistance clusters together with two external isolates.
Conclusions: We report a high rate of MDR and low-level ciprofloxacin resistant S. Typhi circulating in Zanzibar, belonging to genotype 4.3.1.1, which is widespread in Southeast Asia and African countries and associated with low-level ciprofloxacin resistance. Few therapeutic options are available for treatment of typhoid fever in the study setting. Surveillance of the prevalence, spread and antimicrobial susceptibility of S. Typhi can guide treatment and control efforts.
Copyright: © 2024 Onken et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Global Burden of Disease Collaborative Network. GBD 2020 Cause and Risk Summaries: Typhoid fever—level 4 cause. Seattle, United States: Institute for Health Metrics and Evaluation (IHME). 2020. [cited 2022 07.02.2022]. Available from: https://www.healthdata.org/results/gbd_summaries/2019/typhoid-fever-leve....
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

