The GRAS transcription factor CsTL regulates tendril formation in cucumber
- PMID: 38630900
- PMCID: PMC11289639
- DOI: 10.1093/plcell/koae123
The GRAS transcription factor CsTL regulates tendril formation in cucumber
Abstract
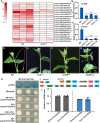
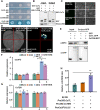
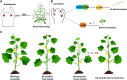
Cucumber (Cucumis sativus, Cs) tendrils are slender vegetative organs that typically require manual removal to ensure orderly growth during greenhouse cultivation. Here, we identified cucumber tendril-less (tl), a Tnt1 retrotransposon-induced insertion mutant lacking tendrils. Map-based cloning identified the mutated gene, CsaV3_3G003590, which we designated as CsTL, which is homologous to Arabidopsis thaliana LATERAL SUPPRESSOR (AtLAS). Knocking out CsTL repressed tendril formation but did not affect branch initiation, whereas overexpression (OE) of CsTL resulted in the formation of two or more tendrils in one leaf axil. Although expression of two cucumber genes regulating tendril formation, Tendril (CsTEN) and Unusual Floral Organs (CsUFO), was significantly decreased in CsTL knockout lines, these two genes were not direct downstream targets of CsTL. Instead, CsTL physically interacted with CsTEN, an interaction that further enhanced CsTEN-mediated expression of CsUFO. In Arabidopsis, the CsTL homolog AtLAS acts upstream of REVOLUTA (REV) to regulate branch initiation. Knocking out cucumber CsREV inhibited branch formation without affecting tendril initiation. Furthermore, genomic regions containing CsTL and AtLAS were not syntenic between the cucumber and Arabidopsis genomes, whereas REV orthologs were found on a shared syntenic block. Our results revealed not only that cucumber CsTL possesses a divergent function in promoting tendril formation but also that CsREV retains its conserved function in shoot branching.
© The Author(s) 2024. Published by Oxford University Press on behalf of American Society of Plant Biologists. All rights reserved. For commercial re-use, please contact reprints@oup.com for reprints and translation rights for reprints. All other permissions can be obtained through our RightsLink service via the Permissions link on the article page on our site—for further information please contact journals.permissions@oup.com.
Conflict of interest statement
Conflict of interest statement. None declared.
Figures




Similar articles
-
Tendril length is determined by gibberellin deactivation during thigmo response in cucumber.Plant J. 2024 Nov;120(3):901-909. doi: 10.1111/tpj.17023. Epub 2024 Sep 11. Plant J. 2024. PMID: 39259667
-
A Rare SNP Identified a TCP Transcription Factor Essential for Tendril Development in Cucumber.Mol Plant. 2015 Dec 7;8(12):1795-808. doi: 10.1016/j.molp.2015.10.005. Epub 2015 Oct 24. Mol Plant. 2015. PMID: 26597500
-
PACLOBUTRAZOL-RESISTANCE4 positively regulates cell expansion to promote tendril elongation in cucumber.Plant Physiol. 2023 Aug 3;192(4):2756-2767. doi: 10.1093/plphys/kiad245. Plant Physiol. 2023. PMID: 37084381
-
CsUFO is involved in the formation of flowers and tendrils in cucumber.Theor Appl Genet. 2021 Jul;134(7):2141-2150. doi: 10.1007/s00122-021-03811-4. Epub 2021 Mar 19. Theor Appl Genet. 2021. PMID: 33740111 Review.
-
A mutation in class III homeodomain-leucine zipper (HD-ZIP III) transcription factor results in curly leaf (cul) in cucumber (Cucumis sativus L.).Theor Appl Genet. 2019 Jan;132(1):113-123. doi: 10.1007/s00122-018-3198-z. Epub 2018 Oct 17. Theor Appl Genet. 2019. PMID: 30334067 Review.
Cited by
-
The quest to find TLC: Tendril-less cucumbers, that is.Plant Cell. 2024 Jul 31;36(8):2751-2752. doi: 10.1093/plcell/koae137. Plant Cell. 2024. PMID: 38693786 Free PMC article. No abstract available.
-
LzSCL9, a Novel GRAS Transcription Factor in Lanzhou Lily (Lilium davidii var. unicolor), Participates in Regulation of Trichokonins-Primed Heat Stress Tolerance.Plants (Basel). 2024 Aug 21;13(16):2330. doi: 10.3390/plants13162330. Plants (Basel). 2024. PMID: 39204766 Free PMC article.
-
Harbingers of bloom: Identifying the molecular mechanisms controlling fruit bloom formation in cucumbers.Plant Cell. 2025 Aug 4;37(8):koaf187. doi: 10.1093/plcell/koaf187. Plant Cell. 2025. PMID: 40720564 Free PMC article. No abstract available.
References
-
- Bellaire A, Ischebeck T, Staedler Y, Weinhaeuser I, Mair A, Parameswaran S, Ito T, Schönenberger J, Weckwerth W. Metabolism and development - integration of micro computed tomography data and metabolite profiling reveals metabolic reprogramming from floral initiation to silique development. New Phytol. 2014:202(1):322–335. 10.1111/nph.12631 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

