Efficient cytometry analysis with FlowSOM in Python boosts interoperability with other single-cell tools
- PMID: 38632080
- PMCID: PMC11052654
- DOI: 10.1093/bioinformatics/btae179
Efficient cytometry analysis with FlowSOM in Python boosts interoperability with other single-cell tools
Abstract
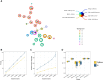
Motivation: We describe a new Python implementation of FlowSOM, a clustering method for cytometry data.
Results: This implementation is faster than the original version in R, better adapted to work with single-cell omics data including integration with current single-cell data structures and includes all the original visualizations, such as the star and pie plot.
Availability and implementation: The FlowSOM Python implementation is freely available on GitHub: https://github.com/saeyslab/FlowSOM_Python.
© The Author(s) 2024. Published by Oxford University Press.
Conflict of interest statement
None declared.
Figures
Similar articles
-
BL-FlowSOM: Consistent and Highly Accelerated FlowSOM Based on Parallelized Batch Learning.Cytometry A. 2025 May;107(5):333-343. doi: 10.1002/cyto.a.24934. Epub 2025 Apr 17. Cytometry A. 2025. PMID: 40243114
-
FlowSOM: Using self-organizing maps for visualization and interpretation of cytometry data.Cytometry A. 2015 Jul;87(7):636-45. doi: 10.1002/cyto.a.22625. Epub 2015 Jan 8. Cytometry A. 2015. PMID: 25573116
-
Analyzing high-dimensional cytometry data using FlowSOM.Nat Protoc. 2021 Aug;16(8):3775-3801. doi: 10.1038/s41596-021-00550-0. Epub 2021 Jun 25. Nat Protoc. 2021. PMID: 34172973 Review.
-
Integration, exploration, and analysis of high-dimensional single-cell cytometry data using Spectre.Cytometry A. 2022 Mar;101(3):237-253. doi: 10.1002/cyto.a.24350. Epub 2021 Apr 26. Cytometry A. 2022. PMID: 33840138
-
Unsupervised flow cytometry analysis in hematological malignancies: A new paradigm.Int J Lab Hematol. 2021 Jul;43 Suppl 1:54-64. doi: 10.1111/ijlh.13548. Int J Lab Hematol. 2021. PMID: 34288436 Review.
Cited by
-
Development of a Spectral Flow Cytometry Analysis Pipeline for High-dimensional Immune Cell Characterization.J Immunol. 2024 Dec 1;213(11):1713-1724. doi: 10.4049/jimmunol.2400370. J Immunol. 2024. PMID: 39451039
-
Computational assessment of measurable residual disease in acute myeloid leukemia using mixture models.Commun Med (Lond). 2024 Dec 19;4(1):271. doi: 10.1038/s43856-024-00700-x. Commun Med (Lond). 2024. PMID: 39702555 Free PMC article.
References
-
- Buttner M, Hempel F, Ryborz T. et al. Pytometry: Flow and Mass Cytometry Analytics in Python. 2022. 10.1101/2022.10.10.511546. - DOI

