Characterization of mutations in hepatitis B virus DNA isolated from Japanese HBsAg-positive blood donors in 2021 and 2022
- PMID: 38632180
- PMCID: PMC11023964
- DOI: 10.1007/s00705-024-06016-4
Characterization of mutations in hepatitis B virus DNA isolated from Japanese HBsAg-positive blood donors in 2021 and 2022
Abstract
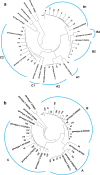
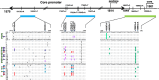
Missense mutations in certain small envelope proteins diminish the efficacy of antibodies. Consequently, tracking the incidence and types of vaccine-escape mutations (VEMs) was crucial both before and after the introduction of universal hepatitis B vaccination in Japan in 2016. In this study, we isolated hepatitis B virus (HBV) DNA from 58 of 169 hepatitis B surface antigen (HBsAg)-positive blood samples from Japanese blood donors and determined the nucleotide sequence encoding the small envelope protein. DNA from six (10%) of the samples had VEMs, but no missense mutations, such as G145R, were detected. Complete HBV genome sequences were obtained from 29 of the 58 samples; the viral genotype was A1 in one (3%), A2 in three (10%), B1 in nine (31%), B2 in five (17%), B4 in one (3%), and C2 in 10 (34%) samples. Tenofovir-resistance mutations were detected in two (7%) samples. In addition, several core promoter mutations, such as 1762A>T and 1764G>A, and a precore nonsense mutation, 1986G>A, which are risk factors for HBV-related chronic liver disease, were detected. These findings provide a baseline for future research and highlight the importance of ongoing monitoring of VEMs and drug resistance mutations in HBV DNA from HBsAg-positive blood donors without HBV antibodies.
© 2024. The Author(s).
Conflict of interest statement
The authors have no relevant financial or non-financial interests to disclose.
Figures





Similar articles
-
Characterization of genetic mutations in hepatitis B virus isolated from HBsAg+/HBcAb+/HBsAb-/HBV DNA + Japanese blood donors.Sci Rep. 2025 Aug 25;15(1):31265. doi: 10.1038/s41598-025-17245-1. Sci Rep. 2025. PMID: 40855119 Free PMC article.
-
Substantial variation in the hepatitis B surface antigen (HBsAg) in hepatitis B virus (HBV)-positive patients from South Africa: Reliable detection of HBV by the Elecsys HBsAg II assay.J Clin Virol. 2018 Apr;101:38-43. doi: 10.1016/j.jcv.2018.01.011. Epub 2018 Feb 6. J Clin Virol. 2018. PMID: 29414186
-
Molecular and serological characterization of hepatitis B virus genotype A and D infected blood donors in Poland.J Viral Hepat. 2010 Jun;17(6):444-52. doi: 10.1111/j.1365-2893.2009.01192.x. Epub 2009 Sep 25. J Viral Hepat. 2010. PMID: 19780948
-
Optimization of the algorithm diagnosis chronic hepatitis B markers in patients with newly diagnosed HIV infection.Klin Lab Diagn. 2020 Sep 16;65(9):574-579. doi: 10.18821/0869-2084-2020-65-9-574-579. Klin Lab Diagn. 2020. PMID: 33245644 English.
-
High-Frequency Notable HBV Mutations Identified in Blood Donors With Occult Hepatitis B Infection From Heyuan City of Southern China.Front Immunol. 2022 May 13;13:754383. doi: 10.3389/fimmu.2022.754383. eCollection 2022. Front Immunol. 2022. PMID: 35634299 Free PMC article.
Cited by
-
An In-Depth Approach to the Associations between MicroRNAs and Viral Load in Patients with Chronic Hepatitis B-A Systematic Review and Meta-Analysis.Int J Mol Sci. 2024 Aug 1;25(15):8410. doi: 10.3390/ijms25158410. Int J Mol Sci. 2024. PMID: 39125978 Free PMC article.
-
Characterization of genetic mutations in hepatitis B virus isolated from HBsAg+/HBcAb+/HBsAb-/HBV DNA + Japanese blood donors.Sci Rep. 2025 Aug 25;15(1):31265. doi: 10.1038/s41598-025-17245-1. Sci Rep. 2025. PMID: 40855119 Free PMC article.
References
-
- Tanaka J, Akita T, Ko K, Miura Y, Satake M; Epidemiological Research Group on Viral Hepatitis and its Long-term Course, Ministry of Health, Labour and Welfare of Japan Countermeasures against viral hepatitis B and C in Japan: an epidemiological point of view. Hepatol Res. 2019;49:990–1002. doi: 10.1111/hepr.13417. - DOI - PMC - PubMed
-
- Tanaka A, Yamagishi N, Hasegawa T, Miyakawa K, Goto N, Matsubayashi K, Satake M. Marked reduction in the incidence of transfusion-transmitted hepatitis B virus infection after the introduction of antibody to hepatitis B core antigen and individual donation nucleic acid amplification screening in Japan. Transfusion. 2023;63:2083–2097. doi: 10.1111/trf.17546. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

