TAK1 inhibition leads to RIPK1-dependent apoptosis in immune-activated cancers
- PMID: 38632238
- PMCID: PMC11024179
- DOI: 10.1038/s41419-024-06654-1
TAK1 inhibition leads to RIPK1-dependent apoptosis in immune-activated cancers
Abstract
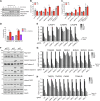
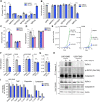
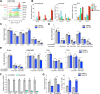
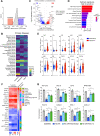
Poor survival and lack of treatment response in glioblastoma (GBM) is attributed to the persistence of glioma stem cells (GSCs). To identify novel therapeutic approaches, we performed CRISPR/Cas9 knockout screens and discovered TGFβ activated kinase (TAK1) as a selective survival factor in a significant fraction of GSCs. Loss of TAK1 kinase activity results in RIPK1-dependent apoptosis via Caspase-8/FADD complex activation, dependent on autocrine TNFα ligand production and constitutive TNFR signaling. We identify a transcriptional signature associated with immune activation and the mesenchymal GBM subtype to be a characteristic of cancer cells sensitive to TAK1 perturbation and employ this signature to accurately predict sensitivity to the TAK1 kinase inhibitor HS-276. In addition, exposure to pro-inflammatory cytokines IFNγ and TNFα can sensitize resistant GSCs to TAK1 inhibition. Our findings reveal dependency on TAK1 kinase activity as a novel vulnerability in immune-activated cancers, including mesenchymal GBMs that can be exploited therapeutically.
© 2024. The Author(s).
Conflict of interest statement
TH, PFH, and SS are co-founders of Eydisbio Inc, a startup company that has licensed the patents related to the HS-276 molecule, used in this study. SP is a co-founder and scientific advisor to Cellinta Ltd. KH is a co-founder of Dania Therapeutics, and a scientific advisor to Hannibal Innovation. BS is now employed as a Senior Bioinformatician at Zifo RnD solutions. TH, PFH, and SS are co-inventors of three US patent filings related to the HS-276 scaffold. The remaining authors declare no competing interests.
Figures



References
-
- Wen PY, Weller M, Lee EQ, Alexander BM, Barnholtz-Sloan JS, Barthel FP, et al. Glioblastoma in adults: a Society for Neuro-Oncology (SNO) and European Society of Neuro-Oncology (EANO) consensus review on current management and future directions. Neuro Oncol. 2020;22:1073–113. doi: 10.1093/neuonc/noaa106. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

