Non-animal models for blood-brain barrier permeability evaluation of drug-like compounds
- PMID: 38632344
- PMCID: PMC11024088
- DOI: 10.1038/s41598-024-59734-9
Non-animal models for blood-brain barrier permeability evaluation of drug-like compounds
Abstract
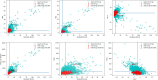
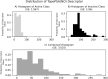
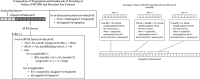
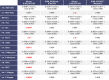
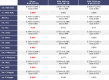
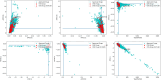
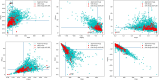
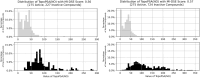
Diseases related to the central nervous system (CNS) are major health concerns and have serious social and economic impacts. Developing new drugs for CNS-related disorders presents a major challenge as it actively involves delivering drugs into the CNS. Therefore, it is imperative to develop in silico methodologies to reliably identify potential lead compounds that can penetrate the blood-brain barrier (BBB) and help to thoroughly understand the role of different physicochemical properties fundamental to the BBB permeation of molecules. In this study, we have analysed the chemical space of the CNS drugs and compared it to the non-CNS-approved drugs. Additionally, we have collected a feature selection dataset from Muehlbacher et al. (J Comput Aided Mol Des 25(12):1095-1106, 2011. 10.1007/s10822-011-9478-1) and an in-house dataset. This information was utilised to design a molecular fingerprint that was used to train machine learning (ML) models. The best-performing models reported in this study achieved accuracies of 0.997 and 0.98, sensitivities of 1.0 and 0.992, specificities of 0.971 and 0.962, MCCs of 0.984 and 0.958, and ROC-AUCs of 0.997 and 0.999 on an imbalanced and a balanced dataset, respectively. They demonstrated overall good accuracies and sensitivities in the blind validation dataset. The reported models can be applied for fast and early screening drug-like molecules with BBB potential. Furthermore, the bbbPythoN package can be used by the research community to both produce the BBB-specific molecular fingerprints and employ the models mentioned earlier for BBB-permeability prediction.
Keywords: Blood–brain barrier; CNS; CNS drug discovery; Central nervous system; Computational prediction; Machine learning; Model; Permeability.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Machine Learning in Drug Development for Neurological Diseases: A Review of Blood Brain Barrier Permeability Prediction Models.Mol Inform. 2025 Mar;44(3):e202400325. doi: 10.1002/minf.202400325. Mol Inform. 2025. PMID: 40146590 Free PMC article. Review.
-
Uncovering blood-brain barrier permeability: a comparative study of machine learning models using molecular fingerprints, and SHAP explainability.SAR QSAR Environ Res. 2024 Dec;35(12):1155-1171. doi: 10.1080/1062936X.2024.2446352. Epub 2025 Jan 8. SAR QSAR Environ Res. 2024. PMID: 39773123
-
Prediction of the Blood-Brain Barrier (BBB) Permeability of Chemicals Based on Machine-Learning and Ensemble Methods.Chem Res Toxicol. 2021 Jun 21;34(6):1456-1467. doi: 10.1021/acs.chemrestox.0c00343. Epub 2021 May 28. Chem Res Toxicol. 2021. PMID: 34047182
-
Predicting blood-brain barrier permeability of molecules with a large language model and machine learning.Sci Rep. 2024 Jul 9;14(1):15844. doi: 10.1038/s41598-024-66897-y. Sci Rep. 2024. PMID: 38982309 Free PMC article.
-
Blood-brain barrier structure and function and the challenges for CNS drug delivery.J Inherit Metab Dis. 2013 May;36(3):437-49. doi: 10.1007/s10545-013-9608-0. Epub 2013 Apr 23. J Inherit Metab Dis. 2013. PMID: 23609350 Review.
Cited by
-
Development of a succinyl CoA:3-ketoacid CoA transferase inhibitor selective for peripheral tissues that improves glycemia in obesity.iScience. 2025 Apr 3;28(5):112336. doi: 10.1016/j.isci.2025.112336. eCollection 2025 May 16. iScience. 2025. PMID: 40454095 Free PMC article.
-
Machine Learning in Drug Development for Neurological Diseases: A Review of Blood Brain Barrier Permeability Prediction Models.Mol Inform. 2025 Mar;44(3):e202400325. doi: 10.1002/minf.202400325. Mol Inform. 2025. PMID: 40146590 Free PMC article. Review.
-
Artificial intelligence-driven prediction and validation of blood-brain barrier permeability and absorption, distribution, metabolism, excretion profiles in natural product research laboratory compounds.Biomedicine (Taipei). 2024 Dec 1;14(4):82-91. doi: 10.37796/2211-8039.1474. eCollection 2024. Biomedicine (Taipei). 2024. PMID: 39777113 Free PMC article.
References
MeSH terms
LinkOut - more resources
Full Text Sources

