Shark genome size evolution and its relationship with cellular, life-history, ecological, and diversity traits
- PMID: 38632352
- PMCID: PMC11024215
- DOI: 10.1038/s41598-024-59202-4
Shark genome size evolution and its relationship with cellular, life-history, ecological, and diversity traits
Abstract
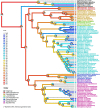
Among vertebrates, sharks exhibit both large and heterogeneous genome sizes ranging from 2.86 to 17.05 pg. Aiming for a better understanding of the patterns and causalities of shark genome size evolution, we applied phylogenetic comparative methods to published genome-size estimates for 71 species representing the main phylogenetic lineages, life-histories and ecological traits. The sixfold range of genome size variation was strongly traceable throughout the phylogeny, with a major expansion preceding shark diversification during the late Paleozoic and an ancestral state (6.33 pg) close to the present-day average (6.72 pg). Subsequent deviations from this average occurred at higher rates in squalomorph than in galeomorph sharks and were unconnected to evolutionary changes in the karyotype architecture, which were dominated by descending disploidy events. Genome size was positively correlated with cell and nucleus sizes and negatively with metabolic rate. The metabolic constraints on increasing genome size also manifested at higher phenotypic scales, with large genomes associated with slow lifestyles and purely marine waters. Moreover, large genome sizes were also linked to non-placental reproductive modes, which may entail metabolically less demanding embryological developments. Contrary to ray-finned fishes, large genome size was associated neither with the taxonomic diversity of affected clades nor with low genetic diversity.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Klug S. Monophyly, phylogeny and systematic position of the †Synechodontiformes (Chondrichthyes, Neoselachii) Zool. Scr. 2009;39:37–49. doi: 10.1111/j.1463-6409.2009.00399.x. - DOI
-
- Pollerspöck, J. & Straube, N. Bibliography database of living/fossil sharks, rays and chimaeras (Chondrichthyes: Elasmobranchii, Holocephali)—List of valid extant species; list of described extant species; statistic. Version 08/2023. World Wide Web Electronic Publication. www.shark-references.com (2023).
-
- Pearce J, Fraser MW, Sequeira AMM, Kaur P. State of shark and ray genomics in an era of extinction. Front. Mar. Sci. 2021;8:415–421. doi: 10.3389/fmars.2021.744986. - DOI
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

