The dual role of Spn-E in supporting heterotypic ping-pong piRNA amplification in silkworms
- PMID: 38632376
- PMCID: PMC11094040
- DOI: 10.1038/s44319-024-00137-2
The dual role of Spn-E in supporting heterotypic ping-pong piRNA amplification in silkworms
Abstract
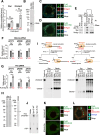
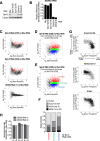
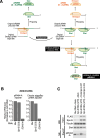
The PIWI-interacting RNA (piRNA) pathway plays a crucial role in silencing transposons in the germline. piRNA-guided target cleavage by PIWI proteins triggers the biogenesis of new piRNAs from the cleaved RNA fragments. This process, known as the ping-pong cycle, is mediated by the two PIWI proteins, Siwi and BmAgo3, in silkworms. However, the detailed molecular mechanism of the ping-pong cycle remains largely unclear. Here, we show that Spindle-E (Spn-E), a putative ATP-dependent RNA helicase, is essential for BmAgo3-dependent production of Siwi-bound piRNAs in the ping-pong cycle and that this function of Spn-E requires its ATPase activity. Moreover, Spn-E acts to suppress homotypic Siwi-Siwi ping-pong, but this function of Spn-E is independent of its ATPase activity. These results highlight the dual role of Spn-E in facilitating proper heterotypic ping-pong in silkworms.
Keywords: BmAgo3; Ping-Pong Cycle; Siwi; Spn-E; piRNA.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
RNA helicase Spn-E is required to maintain Aub and AGO3 protein levels for piRNA silencing in the germline of Drosophila.Eur J Cell Biol. 2016 Sep;95(9):311-22. doi: 10.1016/j.ejcb.2016.06.001. Epub 2016 Jun 6. Eur J Cell Biol. 2016. PMID: 27320195
-
Respective functions of two distinct Siwi complexes assembled during PIWI-interacting RNA biogenesis in Bombyx germ cells.Cell Rep. 2015 Jan 13;10(2):193-203. doi: 10.1016/j.celrep.2014.12.013. Epub 2014 Dec 31. Cell Rep. 2015. PMID: 25558067
-
[The interplay of transposon silencing genes in the Drosophila melanogaster germline].Mol Biol (Mosk). 2011 Jul-Aug;45(4):633-41. Mol Biol (Mosk). 2011. PMID: 21954595 Russian.
-
One Loop to Rule Them All: The Ping-Pong Cycle and piRNA-Guided Silencing.Trends Biochem Sci. 2016 Apr;41(4):324-337. doi: 10.1016/j.tibs.2015.12.008. Epub 2016 Jan 19. Trends Biochem Sci. 2016. PMID: 26810602 Free PMC article. Review.
-
piRNA-Guided Genome Defense: From Biogenesis to Silencing.Annu Rev Genet. 2018 Nov 23;52:131-157. doi: 10.1146/annurev-genet-120417-031441. Annu Rev Genet. 2018. PMID: 30476449 Free PMC article. Review.
Cited by
-
A conserved PIWI silencing complex detects piRNA-target engagement.Mol Cell. 2025 Sep 4;85(17):3275-3287.e7. doi: 10.1016/j.molcel.2025.08.010. Mol Cell. 2025. PMID: 40912244
-
piRNA processing within non-membrane structures is governed by constituent proteins and their functional motifs.FEBS J. 2025 Jun;292(11):2715-2736. doi: 10.1111/febs.17360. Epub 2024 Dec 30. FEBS J. 2025. PMID: 39739617 Free PMC article. Review.
-
A model that integrates the different piRNA biogenesis pathways based on studies in silkworm BmN4 cells.Curr Res Insect Sci. 2025 Feb 11;7:100108. doi: 10.1016/j.cris.2025.100108. eCollection 2025. Curr Res Insect Sci. 2025. PMID: 40083348 Free PMC article. Review.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

