Comprehensive genomic analysis of the SARS-CoV-2 Omicron variant BA.2.76 in Jining City, China, 2022
- PMID: 38632523
- PMCID: PMC11022347
- DOI: 10.1186/s12864-024-10246-w
Comprehensive genomic analysis of the SARS-CoV-2 Omicron variant BA.2.76 in Jining City, China, 2022
Abstract
Objective: This study aims to analyze the molecular characteristics of the novel coronavirus (SARS-CoV-2) Omicron variant BA.2.76 in Jining City, China.
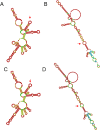
Methods: Whole-genome sequencing was performed on 87 cases of SARS-CoV-2 infection. Evolutionary trees were constructed using bioinformatics software to analyze sequence homology, variant sites, N-glycosylation sites, and phosphorylation sites.
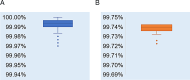
Results: All 87 SARS-CoV-2 whole-genome sequences were classified under the evolutionary branch of the Omicron variant BA.2.76. Their similarity to the reference strain Wuhan-Hu-1 ranged from 99.72 to 99.74%. In comparison to the reference strain Wuhan-Hu-1, the 87 sequences exhibited 77-84 nucleotide differences and 27 nucleotide deletions. A total of 69 amino acid variant sites, 9 amino acid deletions, and 1 stop codon mutation were identified across 18 proteins. Among them, the spike (S) protein exhibited the highest number of variant sites, and the ORF8 protein showed a Q27 stop mutation. Multiple proteins displayed variations in glycosylation and phosphorylation sites.
Conclusion: SARS-CoV-2 continues to evolve, giving rise to new strains with enhanced transmission, stronger immune evasion capabilities, and reduced pathogenicity. The application of high-throughput sequencing technologies in the epidemic prevention and control of COVID-19 provides crucial insights into the evolutionary and variant characteristics of the virus at the genomic level, thereby holding significant implications for the prevention and control of the COVID-19 pandemic.
Keywords: Molecular features; Omicron; SARS-CoV-2; Termination mutation; Whole genome sequencing.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Similar articles
-
A comprehensive genomic study, mutation screening, phylogenetic and statistical analysis of SARS-CoV-2 and its variant omicron among different countries.J Infect Public Health. 2022 Aug;15(8):878-891. doi: 10.1016/j.jiph.2022.07.002. Epub 2022 Jul 8. J Infect Public Health. 2022. PMID: 35839568 Free PMC article.
-
Unraveling the Dynamics of Omicron (BA.1, BA.2, and BA.5) Waves and Emergence of the Deltacton Variant: Genomic Epidemiology of the SARS-CoV-2 Epidemic in Cyprus (Oct 2021-Oct 2022).Viruses. 2023 Sep 15;15(9):1933. doi: 10.3390/v15091933. Viruses. 2023. PMID: 37766339 Free PMC article.
-
In silico evaluation of the impact of Omicron variant of concern sublineage BA.4 and BA.5 on the sensitivity of RT-qPCR assays for SARS-CoV-2 detection using whole genome sequencing.J Med Virol. 2023 Jan;95(1):e28241. doi: 10.1002/jmv.28241. Epub 2022 Nov 8. J Med Virol. 2023. PMID: 36263448 Free PMC article.
-
Molecular evolution of SARS-CoV-2 from December 2019 to August 2022.J Med Virol. 2023 Jan;95(1):e28366. doi: 10.1002/jmv.28366. J Med Virol. 2023. PMID: 36458547 Free PMC article. Review.
-
Omicron variant in COVID-19 current pandemic: a reason for apprehension.Horm Mol Biol Clin Investig. 2022 Sep 5;44(1):89-96. doi: 10.1515/hmbci-2022-0010. eCollection 2023 Mar 1. Horm Mol Biol Clin Investig. 2022. PMID: 36064193 Review.
Cited by
-
Effectiveness and Safety of Simnotrelvir/Ritonavir and Nirmatrelvir/Ritonavir in the Treatment of Moderate to Severe COVID-19.Immun Inflamm Dis. 2025 Apr;13(4):e70174. doi: 10.1002/iid3.70174. Immun Inflamm Dis. 2025. PMID: 40226977 Free PMC article.
-
Epidemiological Characteristics and Genotypic Features of Rotavirus and Norovirus in Jining City, 2021-2022.Viruses. 2024 Jun 7;16(6):925. doi: 10.3390/v16060925. Viruses. 2024. PMID: 38932216 Free PMC article.
-
Molecular analysis of 4/91-like variants of avian infectious bronchitis virus (IBV) obtained after the introduction of a 4/91 live-attenuated vaccine in Costa Rica during 2017.Virusdisease. 2025 Mar;36(1):81-92. doi: 10.1007/s13337-025-00910-4. Epub 2025 Feb 28. Virusdisease. 2025. PMID: 40290772
-
Epidemiological characteristics and genomic analysis of respiratory adenovirus in Jining City from February 2023 to July 2024.BMC Genomics. 2025 Apr 14;26(1):369. doi: 10.1186/s12864-025-11558-1. BMC Genomics. 2025. PMID: 40229673 Free PMC article.
References
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous