Respiratory carriage of hypervirulent Klebsiella pneumoniae by indigenous populations of Malaysia
- PMID: 38632538
- PMCID: PMC11025145
- DOI: 10.1186/s12864-024-10276-4
Respiratory carriage of hypervirulent Klebsiella pneumoniae by indigenous populations of Malaysia
Abstract
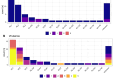
Klebsiella pneumoniae is a Gram-negative Enterobacteriaceae that is classified by the World Health Organisation (WHO) as a Priority One ESKAPE pathogen. South and Southeast Asian countries are regions where both healthcare associated infections (HAI) and community acquired infections (CAI) due to extended-spectrum β-lactamase (ESBL)-producing and carbapenem-resistant K. pneumoniae (CRKp) are of concern. As K. pneumoniae can also exist as a harmless commensal, the spread of resistance genotypes requires epidemiological vigilance. However there has been no significant study of carriage isolates from healthy individuals, particularly in Southeast Asia, and specially Malaysia. Here we describe the genomic analysis of respiratory isolates of K. pneumoniae obtained from Orang Ulu and Orang Asli communities in Malaysian Borneo and Peninsular Malaysia respectively. The majority of isolates were K. pneumoniae species complex (KpSC) 1 K. pneumoniae (n = 53, 89.8%). Four Klebsiella variicola subsp. variicola (KpSC3) and two Klebsiella quasipneumoniae subsp. similipneumoniae (KpSC4) were also found. It was discovered that 30.2% (n = 16) of the KpSC1 isolates were ST23, 11.3% (n = 6) were of ST65, 7.5% (n = 4) were ST13, and 13.2% (n = 7) were ST86. Only eight of the KpSC1 isolates encoded ESBL, but importantly not carbapenemase. Thirteen of the KpSC1 isolates carried yersiniabactin, colibactin and aerobactin, all of which harboured the rmpADC locus and are therefore characterised as hypervirulent. Co-carriage of multiple strains was minimal. In conclusion, most isolates were KpSC1, ST23, one of the most common sequence types and previously found in cases of K. pneumoniae infection. A proportion were hypervirulent (hvKp) however antibiotic resistance was low.
Keywords: Klebsiella pneumoniae; Antimicrobial resistance; ESBL; Hypervirulent; Malaysia.
© 2024. The Author(s).
Conflict of interest statement
DWC was a post-doctoral researcher on projects funded by Pfizer and GSK between April 2014 and October 2017. SCC acts as principal investigator on studies conducted on behalf of University Hospital Southampton NHS Foundation Trust/University of Southampton that are sponsored by vaccine manufacturers but receives no personal payments from them. SCC has participated in advisory boards for vaccine manufacturers but receives no personal payments for this work. SCC has received financial assistance from vaccine manufacturers to attend conferences. All grants and honoraria are paid into accounts within the respective NHS Trusts or Universities, or to independent charities. All other authors have no conflicts of interest.
Figures



References
-
- Carville KS, Lehmann D, Hall G, Moore H, Richmond P, de Klerk N et al. Infection is the major component of the Disease Burden in Aboriginal and non-aboriginal Australian children: a Population-based study. Pediatr Infect Dis J. 2007;26(3). - PubMed
-
- SyedHussain T, Krishnasamy DS, Hassan AAG. Distribution and demography of the Orang Asli in Malaysia. Int J Humanit Social Sci Invention. 2017;6(1):40–5.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

