Development of a Trusted Third Party at a Large University Hospital: Design and Implementation Study
- PMID: 38632712
- PMCID: PMC11040164
- DOI: 10.2196/53075
Development of a Trusted Third Party at a Large University Hospital: Design and Implementation Study
Abstract
Background: Pseudonymization has become a best practice to securely manage the identities of patients and study participants in medical research projects and data sharing initiatives. This method offers the advantage of not requiring the direct identification of data to support various research processes while still allowing for advanced processing activities, such as data linkage. Often, pseudonymization and related functionalities are bundled in specific technical and organization units known as trusted third parties (TTPs). However, pseudonymization can significantly increase the complexity of data management and research workflows, necessitating adequate tool support. Common tasks of TTPs include supporting the secure registration and pseudonymization of patient and sample identities as well as managing consent.
Objective: Despite the challenges involved, little has been published about successful architectures and functional tools for implementing TTPs in large university hospitals. The aim of this paper is to fill this research gap by describing the software architecture and tool set developed and deployed as part of a TTP established at Charité - Universitätsmedizin Berlin.
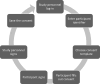
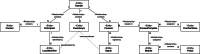
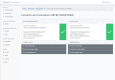
Methods: The infrastructure for the TTP was designed to provide a modular structure while keeping maintenance requirements low. Basic functionalities were realized with the free MOSAIC tools. However, supporting common study processes requires implementing workflows that span different basic services, such as patient registration, followed by pseudonym generation and concluded by consent collection. To achieve this, an integration layer was developed to provide a unified Representational state transfer (REST) application programming interface (API) as a basis for more complex workflows. Based on this API, a unified graphical user interface was also implemented, providing an integrated view of information objects and workflows supported by the TTP. The API was implemented using Java and Spring Boot, while the graphical user interface was implemented in PHP and Laravel. Both services use a shared Keycloak instance as a unified management system for roles and rights.
Results: By the end of 2022, the TTP has already supported more than 10 research projects since its launch in December 2019. Within these projects, more than 3000 identities were stored, more than 30,000 pseudonyms were generated, and more than 1500 consent forms were submitted. In total, more than 150 people regularly work with the software platform. By implementing the integration layer and the unified user interface, together with comprehensive roles and rights management, the effort for operating the TTP could be significantly reduced, as personnel of the supported research projects can use many functionalities independently.
Conclusions: With the architecture and components described, we created a user-friendly and compliant environment for supporting research projects. We believe that the insights into the design and implementation of our TTP can help other institutions to efficiently and effectively set up corresponding structures.
Keywords: EHR; application; architecture; consent; data management; data privacy; electronic health record; health platform; health record; identifying data; implementation; infrastructure; modular; pseudonymisation; pseudonymization; scalability; security; software; trusted third party; user interface.
© Eric Wündisch, Peter Hufnagl, Peter Brunecker, Sophie Meier zu Ummeln, Sarah Träger, Marcus Kopp, Fabian Prasser, Joachim Weber. Originally published in JMIR Medical Informatics (https://medinform.jmir.org).
Conflict of interest statement
Figures
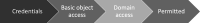




Similar articles
-
The University Medicine Greifswald's Trusted Third Party Dispatcher: State-of-the-Art Perspective Into Comprehensive Architectures and Complex Research Workflows.JMIR Med Inform. 2024 Nov 29;12:e65784. doi: 10.2196/65784. JMIR Med Inform. 2024. PMID: 39612471 Free PMC article. No abstract available.
-
Authors' Reply: The University Medicine Greifswald's Trusted Third Party Dispatcher: State-of-the-Art Perspective Into Comprehensive Architectures and Complex Research Workflows.JMIR Med Inform. 2024 Nov 29;12:e67429. doi: 10.2196/67429. JMIR Med Inform. 2024. PMID: 39612473 Free PMC article. No abstract available.
-
Integration of Trusted Third Party Software into an EDC System for Data Protection - Compliant Identity Management, Consent Management and Pseudonymization in Medical Research Studies.Stud Health Technol Inform. 2024 Aug 30;317:75-84. doi: 10.3233/SHTI240840. Stud Health Technol Inform. 2024. PMID: 39234709
-
Pseudonymization tools for medical research: a systematic review.BMC Med Inform Decis Mak. 2025 Mar 12;25(1):128. doi: 10.1186/s12911-025-02958-0. BMC Med Inform Decis Mak. 2025. PMID: 40075358 Free PMC article.
-
Privacy-Preserving Methods for Feature Engineering Using Blockchain: Review, Evaluation, and Proof of Concept.J Med Internet Res. 2019 Aug 14;21(8):e13600. doi: 10.2196/13600. J Med Internet Res. 2019. PMID: 31414666 Free PMC article. Review.
Cited by
-
The University Medicine Greifswald's Trusted Third Party Dispatcher: State-of-the-Art Perspective Into Comprehensive Architectures and Complex Research Workflows.JMIR Med Inform. 2024 Nov 29;12:e65784. doi: 10.2196/65784. JMIR Med Inform. 2024. PMID: 39612471 Free PMC article. No abstract available.
-
Authors' Reply: The University Medicine Greifswald's Trusted Third Party Dispatcher: State-of-the-Art Perspective Into Comprehensive Architectures and Complex Research Workflows.JMIR Med Inform. 2024 Nov 29;12:e67429. doi: 10.2196/67429. JMIR Med Inform. 2024. PMID: 39612473 Free PMC article. No abstract available.
References
-
- Pommerening K, Sax U, Müller T, Speer R, Ganslandt T, Drepper J. Integrating eHealth and medical research: the TMF data protection scheme. [10-04-2024];EHealth Comb Health Telemat Telemed Biomed Eng Bioinforma Edge. 2008 Jan;:5–10. https://www.staff.uni-mainz.de/pommeren/Artikel/CeHR_POM_Publ.pdf URL. Accessed.
LinkOut - more resources
Full Text Sources
Miscellaneous

