Removing crosstalk signals in neuron activity by time multiplexed excitations in a two-photon all-optical physiology system
- PMID: 38633062
- PMCID: PMC11019693
- DOI: 10.1364/BOE.521047
Removing crosstalk signals in neuron activity by time multiplexed excitations in a two-photon all-optical physiology system
Abstract
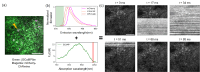
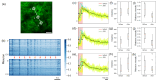
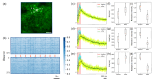
The two-photon all-optical physiology system has attracted great interest in deciphering neuronal circuits in vivo, benefiting from its advantages in recording and modulating neuronal activities at single neuron resolutions. However, the interference, or crosstalk, between the imaging and photostimulation beams introduces a significant challenge and may impede the future application of voltage indicators in two-photon all-optical physiology system. Here, we propose the time multiplexed excitation method to distinguish signals from neuronal activities and crosstalks from photostimulation. In our system, the laser pulses of the imaging beam and photostimulation beam are synchronized, and a time delay is introduced into these pulses to separate the fluorescence signal generated by these two beams. We demonstrate the efficacy of our system in eliminating crosstalk signals from photostimulation and evaluate its influence on both genetically encoded calcium indicators (GECIs) and genetically encoded voltage indicators (GEVIs) through in vivo experiments.
© 2024 Optica Publishing Group.
Conflict of interest statement
The authors declare that there are no conflicts of interest related to this article.
Figures



Similar articles
-
A novel rhodopsin-based voltage indicator for simultaneous two-photon optical recording with GCaMP in vivo.bioRxiv [Preprint]. 2024 Nov 17:2024.11.15.623698. doi: 10.1101/2024.11.15.623698. bioRxiv. 2024. PMID: 39605646 Free PMC article. Preprint.
-
Using Genetically Encoded Voltage Indicators (GEVIs) to Study the Input-Output Transformation of the Mammalian Olfactory Bulb.Front Cell Neurosci. 2019 Jul 31;13:342. doi: 10.3389/fncel.2019.00342. eCollection 2019. Front Cell Neurosci. 2019. PMID: 31417362 Free PMC article.
-
High-speed multiplane confocal microscopy for voltage imaging in densely labeled neuronal populations.Nat Neurosci. 2023 Sep;26(9):1642-1650. doi: 10.1038/s41593-023-01408-2. Epub 2023 Aug 21. Nat Neurosci. 2023. PMID: 37604887 Free PMC article.
-
Toward Better Genetically Encoded Sensors of Membrane Potential.Trends Neurosci. 2016 May;39(5):277-289. doi: 10.1016/j.tins.2016.02.005. Trends Neurosci. 2016. PMID: 27130905 Free PMC article. Review.
-
Voltage imaging to understand connections and functions of neuronal circuits.J Neurophysiol. 2016 Jul 1;116(1):135-52. doi: 10.1152/jn.00226.2016. Epub 2016 Apr 13. J Neurophysiol. 2016. PMID: 27075539 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
