Optical coherence tomography for multicellular tumor spheroid category recognition and drug screening classification via multi-spatial-superficial-parameter and machine learning
- PMID: 38633082
- PMCID: PMC11019711
- DOI: 10.1364/BOE.514079
Optical coherence tomography for multicellular tumor spheroid category recognition and drug screening classification via multi-spatial-superficial-parameter and machine learning
Abstract
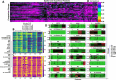
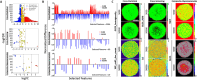
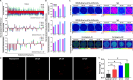
Optical coherence tomography (OCT) is an ideal imaging technique for noninvasive and longitudinal monitoring of multicellular tumor spheroids (MCTS). However, the internal structure features within MCTS from OCT images are still not fully utilized. In this study, we developed cross-statistical, cross-screening, and composite-hyperparameter feature processing methods in conjunction with 12 machine learning models to assess changes within the MCTS internal structure. Our results indicated that the effective features combined with supervised learning models successfully classify OVCAR-8 MCTS culturing with 5,000 and 50,000 cell numbers, MCTS with pancreatic tumor cells (Panc02-H7) culturing with the ratio of 0%, 33%, 50%, and 67% of fibroblasts, and OVCAR-4 MCTS treated by 2-methoxyestradiol, AZD1208, and R-ketorolac with concentrations of 1, 10, and 25 µM. This approach holds promise for obtaining multi-dimensional physiological and functional evaluations for using OCT and MCTS in anticancer studies.
© 2024 Optica Publishing Group.
Conflict of interest statement
The authors declare no competing interests.
Figures








Similar articles
-
Optical Coherence Tomography of Tumor Spheroids Identifies Candidates for Drug Repurposing in Ovarian Cancer.IEEE Trans Biomed Eng. 2023 Jun;70(6):1891-1901. doi: 10.1109/TBME.2022.3231835. Epub 2023 May 19. IEEE Trans Biomed Eng. 2023. PMID: 37015385 Free PMC article.
-
Optical Coherence Tomography Detects Necrotic Regions and Volumetrically Quantifies Multicellular Tumor Spheroids.Cancer Res. 2017 Nov 1;77(21):6011-6020. doi: 10.1158/0008-5472.CAN-17-0821. Epub 2017 Sep 13. Cancer Res. 2017. PMID: 28904062 Free PMC article.
-
Longitudinal Morphological and Physiological Monitoring of Three-dimensional Tumor Spheroids Using Optical Coherence Tomography.J Vis Exp. 2019 Feb 9;(144):10.3791/59020. doi: 10.3791/59020. J Vis Exp. 2019. PMID: 30799861 Free PMC article.
-
Challenges of applying multicellular tumor spheroids in preclinical phase.Cancer Cell Int. 2021 Mar 4;21(1):152. doi: 10.1186/s12935-021-01853-8. Cancer Cell Int. 2021. PMID: 33663530 Free PMC article. Review.
-
Multicellular Tumor Spheroids (MCTS) as a 3D In Vitro Evaluation Tool of Nanoparticles.Small. 2018 Mar;14(13):e1702858. doi: 10.1002/smll.201702858. Epub 2018 Feb 16. Small. 2018. PMID: 29450963 Review.
Cited by
-
Automatic renal carcinoma biopsy guidance using forward-viewing endoscopic optical coherence tomography and deep learning.Commun Eng. 2024 Aug 2;3(1):107. doi: 10.1038/s44172-024-00254-9. Commun Eng. 2024. PMID: 39095532 Free PMC article.
References
-
- Yuhas J. M., Li A. P., Martinez A. O., et al. , “A simplified method for production and growth of multicellular tumor spheroids,” Cancer Res. 37(10), 3639–3643 (1977). - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
