Siamese deep learning video flow cytometry for automatic and label-free clinical cervical cancer cell analysis
- PMID: 38633087
- PMCID: PMC11019674
- DOI: 10.1364/BOE.510022
Siamese deep learning video flow cytometry for automatic and label-free clinical cervical cancer cell analysis
Abstract
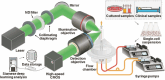
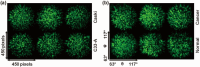
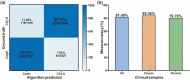
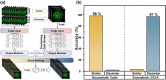
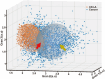
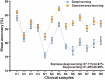
Automatic and label-free screening methods may help to reduce cervical cancer mortality rates, especially in developing regions. The latest advances of deep learning in the biomedical optics field provide a more automatic approach to solving clinical dilemmas. However, existing deep learning methods face challenges, such as the requirement of manually annotated training sets for clinical sample analysis. Here, we develop Siamese deep learning video flow cytometry for the analysis of clinical cervical cancer cell samples in a smear-free manner. High-content light scattering images of label-free single cells are obtained via the video flow cytometer. Siamese deep learning, a self-supervised method, is built to introduce cell lineage cells into an analysis of clinical cells, which utilizes generated similarity metrics as label annotations for clinical cells. Compared with other deep learning methods, Siamese deep learning achieves a higher accuracy of up to 87.11%, with about 5.62% improvement for label-free clinical cervical cancer cell classification. The Siamese deep learning video flow cytometry demonstrated here is promising for automatic, label-free analysis of many types of cells from clinical samples without cell smears.
© 2024 Optica Publishing Group.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Light scattering imaging modal expansion cytometry for label-free single-cell analysis with deep learning.Comput Methods Programs Biomed. 2025 Jun;264:108726. doi: 10.1016/j.cmpb.2025.108726. Epub 2025 Mar 15. Comput Methods Programs Biomed. 2025. PMID: 40112688
-
High-content video flow cytometry with digital cell filtering for label-free cell classification by machine learning.Cytometry A. 2023 Apr;103(4):325-334. doi: 10.1002/cyto.a.24701. Epub 2022 Nov 4. Cytometry A. 2023. PMID: 36287146
-
Differentiating single cervical cells by mitochondrial fluorescence imaging and deep learning-based label-free light scattering with multi-modal static cytometry.Cytometry A. 2023 Mar;103(3):240-250. doi: 10.1002/cyto.a.24684. Epub 2022 Sep 7. Cytometry A. 2023. PMID: 36028474
-
Recent developments in cervical cancer diagnosis using deep learning on whole slide images: An Overview of models, techniques, challenges and future directions.Micron. 2023 Oct;173:103520. doi: 10.1016/j.micron.2023.103520. Epub 2023 Jul 29. Micron. 2023. PMID: 37556898 Review.
-
Towards label-efficient automatic diagnosis and analysis: a comprehensive survey of advanced deep learning-based weakly-supervised, semi-supervised and self-supervised techniques in histopathological image analysis.Phys Med Biol. 2022 Oct 14;67(20). doi: 10.1088/1361-6560/ac910a. Phys Med Biol. 2022. PMID: 36084627 Review.
References
-
- WHO guideline for screening and treatment of cervical pre-cancer lesions for cervical cancer prevention, second edition. Geneva: World Health Organization; 2021. License: CC BY-NC-SA 3.0 IGO. - PubMed
LinkOut - more resources
Full Text Sources
