This is a preprint.
Leveraging large-scale biobank EHRs to enhance pharmacogenetics of cardiometabolic disease medications
- PMID: 38633781
- PMCID: PMC11023668
- DOI: 10.1101/2024.04.06.24305415
Leveraging large-scale biobank EHRs to enhance pharmacogenetics of cardiometabolic disease medications
Update in
-
Leveraging large-scale biobank EHRs to enhance pharmacogenetics of cardiometabolic disease medications.Nat Commun. 2025 Mar 25;16(1):2913. doi: 10.1038/s41467-025-58152-3. Nat Commun. 2025. PMID: 40133288 Free PMC article.
Abstract
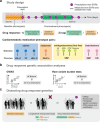
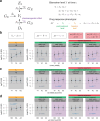
Electronic health records (EHRs) coupled with large-scale biobanks offer great promises to unravel the genetic underpinnings of treatment efficacy. However, medication-induced biomarker trajectories stemming from such records remain poorly studied. Here, we extract clinical and medication prescription data from EHRs and conduct GWAS and rare variant burden tests in the UK Biobank (discovery) and the All of Us program (replication) on ten cardiometabolic drug response outcomes including lipid response to statins, HbA1c response to metformin and blood pressure response to antihypertensives (N = 740-26,669). Our findings at genome-wide significance level recover previously reported pharmacogenetic signals and also include novel associations for lipid response to statins (N = 26,669) near LDLR and ZNF800. Importantly, these associations are treatment-specific and not associated with biomarker progression in medication-naive individuals. Furthermore, we demonstrate that individuals with higher genetically determined low-density and total cholesterol baseline levels experience increased absolute, albeit lower relative biomarker reduction following statin treatment. In summary, we systematically investigated the common and rare pharmacogenetic contribution to cardiometabolic drug response phenotypes in over 50,000 UK Biobank and All of Us participants with EHR and identified clinically relevant genetic predictors for improved personalized treatment strategies.
Conflict of interest statement
Competing interests The authors declare that they have no competing interests.
Figures


References
-
- Nelson Matthew R, Johnson Toby, Warren Liling, Hughes Arlene R, Chissoe Stephanie L, Xu Chun-Fang, and Waterworth Dawn M. The genetics of drug efficacy: opportunities and challenges. Nature Reviews Genetics, 17(4):197–206, 2016. - PubMed
-
- Pirmohamed Munir. Pharmacogenomics: Current status and future perspectives. Nature Reviews Genetics, pages 1–13, 2023. - PubMed
-
- Postmus Iris, Trompet Stella, Deshmukh Harshal A, Barnes Michael R, Li Xiaohui, Warren Helen R, Chasman Daniel I, Zhou Kaixin, Arsenault Benoit J, Donnelly Louise A, et al. Pharmacogenetic meta-analysis of genome-wide association studies of ldl cholesterol response to statins. Nature communications, 5(1):5068, 2014. - PMC - PubMed
-
- Zhou Kaixin, Yee Sook Wah, Seiser Eric L, Van Leeuwen Nienke, Tavendale Roger, Bennett Amanda J, Groves Christopher J, Coleman Ruth L, Van Der Heijden Amber A, Beulens Joline W, et al. Variation in the glucose transporter gene slc2a2 is associated with glycemic response to metformin. Nature genetics, 48(9):1055–1059, 2016. - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
