Development and Validation of an 18-Gene Urine Test for High-Grade Prostate Cancer
- PMID: 38635241
- PMCID: PMC11190811
- DOI: 10.1001/jamaoncol.2024.0455
Development and Validation of an 18-Gene Urine Test for High-Grade Prostate Cancer
Abstract
Importance: Benefits of prostate cancer (PCa) screening with prostate-specific antigen (PSA) alone are largely offset by excess negative biopsies and overdetection of indolent cancers resulting from the poor specificity of PSA for high-grade PCa (ie, grade group [GG] 2 or greater).
Objective: To develop a multiplex urinary panel for high-grade PCa and validate its external performance relative to current guideline-endorsed biomarkers.
Design, setting, and participants: RNA sequencing analysis of 58 724 genes identified 54 markers of PCa, including 17 markers uniquely overexpressed by high-grade cancers. Gene expression and clinical factors were modeled in a new urinary test for high-grade PCa (MyProstateScore 2.0 [MPS2]). Optimal models were developed in parallel without prostate volume (MPS2) and with prostate volume (MPS2+). The locked models underwent blinded external validation in a prospective National Cancer Institute trial cohort. Data were collected from January 2008 to December 2020, and data were analyzed from November 2022 to November 2023.
Exposure: Protocolized blood and urine collection and transrectal ultrasound-guided systematic prostate biopsy.
Main outcomes and measures: Multiple biomarker tests were assessed in the validation cohort, including serum PSA alone, the Prostate Cancer Prevention Trial risk calculator, and the Prostate Health Index (PHI) as well as derived multiplex 2-gene and 3-gene models, the original 2-gene MPS test, and the 18-gene MPS2 models. Under a testing approach with 95% sensitivity for PCa of GG 2 or greater, measures of diagnostic accuracy and clinical consequences of testing were calculated. Cancers of GG 3 or greater were assessed secondarily.
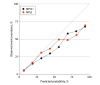
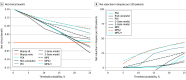
Results: Of 761 men included in the development cohort, the median (IQR) age was 63 (58-68) years, and the median (IQR) PSA level was 5.6 (4.6-7.2) ng/mL; of 743 men included in the validation cohort, the median (IQR) age was 62 (57-68) years, and the median (IQR) PSA level was 5.6 (4.1-8.0) ng/mL. In the validation cohort, 151 (20.3%) had high-grade PCa on biopsy. Area under the receiver operating characteristic curve values were 0.60 using PSA alone, 0.66 using the risk calculator, 0.77 using PHI, 0.76 using the derived multiplex 2-gene model, 0.72 using the derived multiplex 3-gene model, and 0.74 using the original MPS model compared with 0.81 using the MPS2 model and 0.82 using the MPS2+ model. At 95% sensitivity, the MPS2 model would have reduced unnecessary biopsies performed in the initial biopsy population (range for other tests, 15% to 30%; range for MPS2, 35% to 42%) and repeat biopsy population (range for other tests, 9% to 21%; range for MPS2, 46% to 51%). Across pertinent subgroups, the MPS2 models had negative predictive values of 95% to 99% for cancers of GG 2 or greater and of 99% for cancers of GG 3 or greater.
Conclusions and relevance: In this study, a new 18-gene PCa test had higher diagnostic accuracy for high-grade PCa relative to existing biomarker tests. Clinically, use of this test would have meaningfully reduced unnecessary biopsies performed while maintaining highly sensitive detection of high-grade cancers. These data support use of this new PCa biomarker test in patients with elevated PSA levels to reduce the potential harms of PCa screening while preserving its long-term benefits.
Conflict of interest statement
Figures

References
-
- Kocarnik JM, Compton K, Dean FE, et al. ; Global Burden of Disease 2019 Cancer Collaboration . Cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life years for 29 cancer groups from 2010 to 2019: a systematic analysis for the Global Burden of Disease Study 2019. JAMA Oncol. 2022;8(3):420-444. doi:10.1001/jamaoncol.2021.6987 - DOI - PMC - PubMed
-
- National Comprehensive Cancer Network . Prostate cancer early detection (version 1.2023). Accessed May 19, 2023. https://www.nccn.org/professionals/physician_gls/pdf/prostate_detection.pdf - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

