WAV E3 ubiquitin ligases mediate degradation of IAA32/34 in the TMK1-mediated auxin signaling pathway during apical hook development
- PMID: 38635634
- PMCID: PMC11047095
- DOI: 10.1073/pnas.2314353121
WAV E3 ubiquitin ligases mediate degradation of IAA32/34 in the TMK1-mediated auxin signaling pathway during apical hook development
Abstract
Auxin regulates plant growth and development through downstream signaling pathways, including the best-known SCFTIR1/AFB-Aux/IAA-ARF pathway and several other less characterized "noncanonical" pathways. Recently, one SCFTIR1/AFB-independent noncanonical pathway, mediated by Transmembrane Kinase 1 (TMK1), was discovered through the analyses of its functions in Arabidopsis apical hook development. Asymmetric accumulation of auxin on the concave side of the apical hook triggers DAR1-catalyzed release of the C-terminal of TMK1, which migrates into the nucleus, where it phosphorylates and stabilizes IAA32/34 to inhibit cell elongation, which is essential for full apical hook formation. However, the molecular factors mediating IAA32/34 degradation have not been identified. Here, we show that proteins in the CYTOKININ INDUCED ROOT WAVING 1 (CKRW1)/WAVY GROWTH 3 (WAV3) subfamily act as E3 ubiquitin ligases to target IAA32/34 for ubiquitination and degradation, which is inhibited by TMK1c-mediated phosphorylation. This antagonistic interaction between TMK1c and CKRW1/WAV3 subfamily E3 ubiquitin ligases regulates IAA32/34 levels to control differential cell elongation along opposite sides of the apical hook.
Keywords: Aux/IAA proteins; RING-finger E3 ubiquitin ligase; Transmembrane Kinase 1; Wavy Growth 3; noncanonical auxin signaling.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures




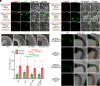
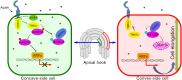
References
-
- Leyser O., Auxin distribution and plant pattern formation: How many angels can dance on the point of PIN? Cell 121, 819–822 (2005). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

