Modeling the emergence of viral resistance for SARS-CoV-2 during treatment with an anti-spike monoclonal antibody
- PMID: 38635853
- PMCID: PMC11060554
- DOI: 10.1371/journal.ppat.1011680
Modeling the emergence of viral resistance for SARS-CoV-2 during treatment with an anti-spike monoclonal antibody
Abstract
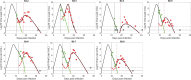
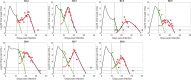
To mitigate the loss of lives during the COVID-19 pandemic, emergency use authorization was given to several anti-SARS-CoV-2 monoclonal antibody (mAb) therapies for the treatment of mild-to-moderate COVID-19 in patients with a high risk of progressing to severe disease. Monoclonal antibodies used to treat SARS-CoV-2 target the spike protein of the virus and block its ability to enter and infect target cells. Monoclonal antibody therapy can thus accelerate the decline in viral load and lower hospitalization rates among high-risk patients with variants susceptible to mAb therapy. However, viral resistance has been observed, in some cases leading to a transient viral rebound that can be as large as 3-4 orders of magnitude. As mAbs represent a proven treatment choice for SARS-CoV-2 and other viral infections, evaluation of treatment-emergent mAb resistance can help uncover underlying pathobiology of SARS-CoV-2 infection and may also help in the development of the next generation of mAb therapies. Although resistance can be expected, the large rebounds observed are much more difficult to explain. We hypothesize replenishment of target cells is necessary to generate the high transient viral rebound. Thus, we formulated two models with different mechanisms for target cell replenishment (homeostatic proliferation and return from an innate immune response antiviral state) and fit them to data from persons with SARS-CoV-2 treated with a mAb. We showed that both models can explain the emergence of resistant virus associated with high transient viral rebounds. We found that variations in the target cell supply rate and adaptive immunity parameters have a strong impact on the magnitude or observability of the viral rebound associated with the emergence of resistant virus. Both variations in target cell supply rate and adaptive immunity parameters may explain why only some individuals develop observable transient resistant viral rebound. Our study highlights the conditions that can lead to resistance and subsequent viral rebound in mAb treatments during acute infection.
Copyright: This is an open access article, free of all copyright, and may be freely reproduced, distributed, transmitted, modified, built upon, or otherwise used by anyone for any lawful purpose. The work is made available under the Creative Commons CC0 public domain dedication.
Conflict of interest statement
We have read the journal’s policy and the authors of this manuscript have the following competing interests: K.W.C. has received research funding to her institution from Merck Sharp & Dohme and consulted for Pardes Biosciences. D.M.S. has consulted for the following companies Fluxergy, Kiadis, Linear Therapies, Matrix BioMed, Lucira, VxBiosciences, Model Medicines, Bayer Pharmaceuticals, and Evidera. E.S.D. has consulted for Gilead, Merck, ViiV and Theratechnologies and received research funding to his institution from Gilead and ViiV. D.A.W. has consulted for Gilead Sciences, ViiV Healthcare, Janssen Pharmaceuticals, and Theratechnologies, and has university grant funding from Gilead Sciences, ViiV Healthcare, and Merck and Co. J. J. E. is on the DMC for Adagio/Invyvid and has consulted for Gilead Sciences and Merck & Co. J. S. C. has consulted for Merck & Co., J.Z.L. has consulted for Abbvie. The other authors have declared that no competing interests exist.
Figures

Update of
-
Modeling the emergence of viral resistance for SARS-CoV-2 during treatment with an anti-spike monoclonal antibody.bioRxiv [Preprint]. 2023 Sep 17:2023.09.14.557679. doi: 10.1101/2023.09.14.557679. bioRxiv. 2023. Update in: PLoS Pathog. 2024 Apr 18;20(4):e1011680. doi: 10.1371/journal.ppat.1011680. PMID: 37745410 Free PMC article. Updated. Preprint.
References
-
- COVID-19 Epidemiological Update ‐ 29 September 2023. Available from: https://www.who.int/publications/m/item/covid-19-epidemiological-update—...
-
- Vaccine equity. Available from: https://www.who.int/campaigns/vaccine-equity
-
- CDC. Coronavirus Disease 2019 (COVID-19). Centers for Disease Control and Prevention. 2020. Available from: https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-classificatio...
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

