CRISPR-Cas9 Screening Identifies KRAS-Induced COX2 as a Driver of Immunotherapy Resistance in Lung Cancer
- PMID: 38635884
- PMCID: PMC11247323
- DOI: 10.1158/0008-5472.CAN-23-2627
CRISPR-Cas9 Screening Identifies KRAS-Induced COX2 as a Driver of Immunotherapy Resistance in Lung Cancer
Abstract
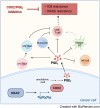
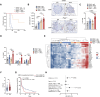
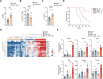
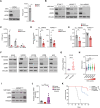
Oncogenic KRAS impairs antitumor immune responses. As effective strategies to combine KRAS inhibitors and immunotherapies have so far proven elusive, a better understanding of the mechanisms by which oncogenic KRAS drives immune evasion is needed to identify approaches that could sensitize KRAS-mutant lung cancer to immunotherapy. In vivo CRISPR-Cas9 screening in an immunogenic murine lung cancer model identified mechanisms by which oncogenic KRAS promotes immune evasion, most notably via upregulation of immunosuppressive COX2 in cancer cells. Oncogenic KRAS potently induced COX2 in both mouse and human lung cancer, which was suppressed using KRAS inhibitors. COX2 acted via prostaglandin E2 (PGE2) to promote resistance to immune checkpoint blockade (ICB) in lung adenocarcinoma. Targeting COX2/PGE2 remodeled the tumor microenvironment by inducing proinflammatory polarization of myeloid cells and influx of activated cytotoxic CD8+ T cells, which increased the efficacy of ICB. Restoration of COX2 expression contributed to tumor relapse after prolonged KRAS inhibition. These results provide the rationale for testing COX2/PGE2 pathway inhibitors in combination with KRASG12C inhibition or ICB in patients with KRAS-mutant lung cancer. Significance: COX2 signaling via prostaglandin E2 is a major mediator of immune evasion driven by oncogenic KRAS that promotes immunotherapy and KRAS-targeted therapy resistance, suggesting effective combination treatments for KRAS-mutant lung cancer.
©2024 The Authors; Published by the American Association for Cancer Research.
Conflict of interest statement
S. de Carné Trécesson reports personal fees from Revolution Medicines outside the submitted work. K. Litchfield reports other support from Isomorphic Labs, personal fees from Monopteros Therapeutics, Ellipses Pharma, Kynos Therapeutics, and SAGA Diagnostics, and grants from Genesis Therapeutics and Cancer ResearchUK/ONO PHARMA/LifeArc IO Alliance outside the submitted work. J. Downward reports grants, personal fees, and nonfinancial support from AstraZeneca, personal fees from Vividion, Jubilant, Theras, and Roche, and grants and nonfinancial support from Bristol Myers Squibb and Revolution Medicines outside the submitted work. No disclosures were reported by the other authors.
Figures



References
-
- Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin 2022;72:7–33. - PubMed
-
- Wei SC, Duffy CR, Allison JP. Fundamental mechanisms of immune checkpoint blockade therapy. Cancer Discov 2018;8:1069–86. - PubMed
-
- Reck M, Rodriguez-Abreu D, Robinson AG, Hui R, Csöszi T, Fülöp A, et al. . Pembrolizumab versus chemotherapy for PD-L1-positive non-small-cell lung cancer. N Engl J Med 2016;375:1823–33. - PubMed
-
- Gandhi L, Rodríguez-Abreu D, Gadgeel S, Esteban E, Felip E, De Angelis F, et al. . Pembrolizumab plus chemotherapy in metastatic non-small-cell lung cancer. N Engl J Med 2018;378:2078–92. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

