Monkeypox virus genomic accordion strategies
- PMID: 38637500
- PMCID: PMC11026394
- DOI: 10.1038/s41467-024-46949-7
Monkeypox virus genomic accordion strategies
Abstract
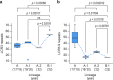
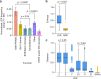
The 2023 monkeypox (mpox) epidemic was caused by a subclade IIb descendant of a monkeypox virus (MPXV) lineage traced back to Nigeria in 1971. Person-to-person transmission appears higher than for clade I or subclade IIa MPXV, possibly caused by genomic changes in subclade IIb MPXV. Key genomic changes could occur in the genome's low-complexity regions (LCRs), which are challenging to sequence and are often dismissed as uninformative. Here, using a combination of highly sensitive techniques, we determine a high-quality MPXV genome sequence of a representative of the current epidemic with LCRs resolved at unprecedented accuracy. This reveals significant variation in short tandem repeats within LCRs. We demonstrate that LCR entropy in the MPXV genome is significantly higher than that of single-nucleotide polymorphisms (SNPs) and that LCRs are not randomly distributed. In silico analyses indicate that expression, translation, stability, or function of MPXV orthologous poxvirus genes (OPGs), including OPG153, OPG204, and OPG208, could be affected in a manner consistent with the established "genomic accordion" evolutionary strategies of orthopoxviruses. We posit that genomic studies focusing on phenotypic MPXV differences should consider LCR variability.
© 2024. The Author(s).
Conflict of interest statement
A.G.-S. has consulting agreements for the following companies involving cash and/or stock: Castlevax, Amovir, Vivaldi Biosciences, Contrafect, 7Hills Pharma, Avimex, Vaxalto, Pagoda, Accurius, Esperovax, Farmak, Applied Biological Laboratories, Pharmamar, Paratus, CureLab Oncology, CureLab Veterinary, Synairgen, and Pfizer, outside of the reported work. A.G.-S. has been an invited speaker in meeting events organized by Seqirus, Janssen, Abbott, and Astrazeneca. A.G.-S. is inventor on patents and patent applications on the use of antivirals and vaccines for the treatment and prevention of virus infections and cancer, owned by the Icahn School of Medicine at Mount Sinai, New York, outside of the reported work. The other authors declare no competing interests.
Figures



References
-
- International Committee on Taxonomy of Viruses. Current ICTV Taxonomy Release. Current ICTV Taxonomy Release. https://ictv.global/taxonomy. (2022).
-
- World Health Organization. ICD-11. International Classification of Diseases 11th Revision. https://icd.who.int/en/. (2022).
-
- Magnus PV, Andersen EK, Petersen KB, Birch-Andersen A. A pox-like disease in cynomolgus monkeys. Acta Pathol. Microbiol. Scand. 2009;46:156–176. doi: 10.1111/j.1699-0463.1959.tb00328.x. - DOI
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

