Immune response stability to the SARS-CoV-2 mRNA vaccine booster is influenced by differential splicing of HLA genes
- PMID: 38637586
- PMCID: PMC11026523
- DOI: 10.1038/s41598-024-59259-1
Immune response stability to the SARS-CoV-2 mRNA vaccine booster is influenced by differential splicing of HLA genes
Abstract
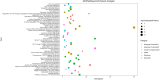
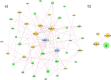
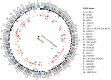
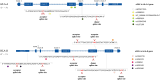
Many molecular mechanisms that lead to the host antibody response to COVID-19 vaccines remain largely unknown. In this study, we used serum antibody detection combined with whole blood RNA-based transcriptome analysis to investigate variability in vaccine response in healthy recipients of a booster (third) dose schedule of the mRNA BNT162b2 vaccine against COVID-19. The cohort was divided into two groups: (1) low-stable individuals, with antibody concentration anti-SARS-CoV IgG S1 below 0.4 percentile at 180 days after boosting vaccination; and (2) high-stable individuals, with antibody values greater than 0.6 percentile of the range in the same period (median 9525 [185-80,000] AU/mL). Differential gene expression, expressed single nucleotide variants and insertions/deletions, differential splicing events, and allelic imbalance were explored to broaden our understanding of the immune response sustenance. Our analysis revealed a differential expression of genes with immunological functions in individuals with low antibody titers, compared to those with higher antibody titers, underscoring the fundamental importance of the innate immune response for boosting immunity. Our findings also provide new insights into the determinants of the immune response variability to the SARS-CoV-2 mRNA vaccine booster, highlighting the significance of differential splicing regulatory mechanisms, mainly concerning HLA alleles, in delineating vaccine immunogenicity.
Keywords: Admixed population; Human leukocyte antigen; Immune response variability; SARS-CoV-2; Vaccine response; Whole blood transcriptome.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Immunogenicity and safety of a booster dose of a self-amplifying RNA COVID-19 vaccine (ARCT-154) versus BNT162b2 mRNA COVID-19 vaccine: a double-blind, multicentre, randomised, controlled, phase 3, non-inferiority trial.Lancet Infect Dis. 2024 Apr;24(4):351-360. doi: 10.1016/S1473-3099(23)00650-3. Epub 2023 Dec 20. Lancet Infect Dis. 2024. PMID: 38141632 Clinical Trial.
-
SARS-CoV-2 specific antibody responses in healthcare workers after a third booster dose of CoronaVac or BNT162b2 vaccine.J Med Virol. 2022 Aug;94(8):3768-3775. doi: 10.1002/jmv.27794. Epub 2022 Apr 23. J Med Virol. 2022. PMID: 35434796 Free PMC article.
-
Evaluation of the safety and immunogenicity of different COVID-19 vaccine combinations in healthy individuals: study protocol for a randomized, subject-blinded, controlled phase 3 trial [PRIBIVAC].Trials. 2022 Jun 16;23(1):498. doi: 10.1186/s13063-022-06345-2. Trials. 2022. PMID: 35710572 Free PMC article.
-
Waning of specific antibodies against Delta and Omicron variants five months after a third dose of BNT162b2 SARS-CoV-2 vaccine in elderly individuals.Front Immunol. 2022 Nov 14;13:1031852. doi: 10.3389/fimmu.2022.1031852. eCollection 2022. Front Immunol. 2022. PMID: 36451833 Free PMC article.
-
Assessment of Neutralizing Antibody Response Against SARS-CoV-2 Variants After 2 to 3 Doses of the BNT162b2 mRNA COVID-19 Vaccine.JAMA Netw Open. 2022 May 2;5(5):e2210780. doi: 10.1001/jamanetworkopen.2022.10780. JAMA Netw Open. 2022. PMID: 35532938 Free PMC article.
Cited by
-
Long-term immune responses to SARS-CoV-2 Omicron BA.4/5 mRNA booster in people living with HIV.Commun Med (Lond). 2025 Mar 27;5(1):92. doi: 10.1038/s43856-025-00799-6. Commun Med (Lond). 2025. PMID: 40148493 Free PMC article.
-
A Multiscale Quantitative Systems Pharmacology Model for the Development and Optimization of mRNA Vaccines.CPT Pharmacometrics Syst Pharmacol. 2025 Jul;14(7):1213-1224. doi: 10.1002/psp4.70041. Epub 2025 May 26. CPT Pharmacometrics Syst Pharmacol. 2025. PMID: 40420402 Free PMC article.
References
MeSH terms
Substances
Grants and funding
- 404096/2020-4/Conselho Nacional de Desenvolvimento Científico e Tecnológico
- 440931/2020-7/Conselho Nacional de Desenvolvimento Científico e Tecnológico
- 302342/2022-2/Conselho Nacional de Desenvolvimento Científico e Tecnológico
- 307145/2021-2/Conselho Nacional de Desenvolvimento Científico e Tecnológico
- 383005/2023-0/Conselho Nacional de Desenvolvimento Científico e Tecnológico
- 310885/2022-1/Conselho Nacional de Desenvolvimento Científico e Tecnológico
- E-26/210.179/2020/Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro
- E-26/211.107/2021/Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro
- E-26/200.883/2021/Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro
- E-26/201.046/2022/Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro
- E-26/202.683/2019/Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro
- E-26/010.00156/2020/Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro
- E-26/211.133/2021/Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro
- E-26/202.376/2021/Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro
- 01.20.0029.000462/20/FINEP
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

