CD20/MS4A1 is a mammalian olfactory receptor expressed in a subset of olfactory sensory neurons that mediates innate avoidance of predators
- PMID: 38637611
- PMCID: PMC11026480
- DOI: 10.1038/s41467-024-47698-3
CD20/MS4A1 is a mammalian olfactory receptor expressed in a subset of olfactory sensory neurons that mediates innate avoidance of predators
Abstract
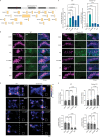
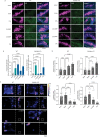
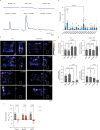
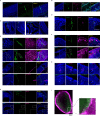
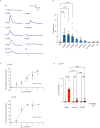
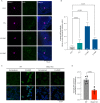
The mammalian olfactory system detects and discriminates between millions of odorants to elicit appropriate behavioral responses. While much has been learned about how olfactory sensory neurons detect odorants and signal their presence, how specific innate, unlearned behaviors are initiated in response to ethologically relevant odors remains poorly understood. Here, we show that the 4-transmembrane protein CD20, also known as MS4A1, is expressed in a previously uncharacterized subpopulation of olfactory sensory neurons in the main olfactory epithelium of the murine nasal cavity and functions as a mammalian olfactory receptor that recognizes compounds produced by mouse predators. While wildtype mice avoid these predator odorants, mice genetically deleted of CD20 do not appropriately respond. Together, this work reveals a CD20-mediated odor-sensing mechanism in the mammalian olfactory system that triggers innate behaviors critical for organismal survival.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
Update of
-
CD20 is a mammalian odorant receptor expressed in a subset of olfactory sensory neurons that mediates innate avoidance of predators.bioRxiv [Preprint]. 2023 Aug 10:2023.08.08.552498. doi: 10.1101/2023.08.08.552498. bioRxiv. 2023. Update in: Nat Commun. 2024 Apr 18;15(1):3360. doi: 10.1038/s41467-024-47698-3. PMID: 37609248 Free PMC article. Updated. Preprint.
-
CD20 is a mammalian odorant receptor expressed in a subset of olfactory sensory neurons that mediates innate avoidance of predators.Res Sq [Preprint]. 2023 Sep 12:rs.3.rs-3290152. doi: 10.21203/rs.3.rs-3290152/v1. Res Sq. 2023. Update in: Nat Commun. 2024 Apr 18;15(1):3360. doi: 10.1038/s41467-024-47698-3. PMID: 37790559 Free PMC article. Updated. Preprint.
Similar articles
-
CD20 is a mammalian odorant receptor expressed in a subset of olfactory sensory neurons that mediates innate avoidance of predators.Res Sq [Preprint]. 2023 Sep 12:rs.3.rs-3290152. doi: 10.21203/rs.3.rs-3290152/v1. Res Sq. 2023. Update in: Nat Commun. 2024 Apr 18;15(1):3360. doi: 10.1038/s41467-024-47698-3. PMID: 37790559 Free PMC article. Updated. Preprint.
-
CD20 is a mammalian odorant receptor expressed in a subset of olfactory sensory neurons that mediates innate avoidance of predators.bioRxiv [Preprint]. 2023 Aug 10:2023.08.08.552498. doi: 10.1101/2023.08.08.552498. bioRxiv. 2023. Update in: Nat Commun. 2024 Apr 18;15(1):3360. doi: 10.1038/s41467-024-47698-3. PMID: 37609248 Free PMC article. Updated. Preprint.
-
Deorphanization of Olfactory Trace Amine-Associated Receptors.Methods Mol Biol. 2025;2915:201-212. doi: 10.1007/978-1-0716-4466-9_14. Methods Mol Biol. 2025. PMID: 40249494
-
Odor discrimination by G protein-coupled olfactory receptors.Microsc Res Tech. 2002 Aug 1;58(3):135-41. doi: 10.1002/jemt.10131. Microsc Res Tech. 2002. PMID: 12203691 Review.
-
Influence of Cat Odor on Reproductive Behavior and Physiology in the House Mouse: (Mus Musculus).In: Mucignat-Caretta C, editor. Neurobiology of Chemical Communication. Boca Raton (FL): CRC Press/Taylor & Francis; 2014. Chapter 14. In: Mucignat-Caretta C, editor. Neurobiology of Chemical Communication. Boca Raton (FL): CRC Press/Taylor & Francis; 2014. Chapter 14. PMID: 24830030 Free Books & Documents. Review.
Cited by
-
MS4A superfamily molecules in tumors, Alzheimer's and autoimmune diseases.Front Immunol. 2024 Dec 9;15:1481494. doi: 10.3389/fimmu.2024.1481494. eCollection 2024. Front Immunol. 2024. PMID: 39717774 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

