Sparassis latifolia and exercise training as complementary medicine mitigated the 5-fluorouracil potent side effects in mice with colorectal cancer: bioinformatics approaches, novel monitoring pathological metrics, screening signatures, and innovative management tactic
- PMID: 38637796
- PMCID: PMC11027426
- DOI: 10.1186/s12935-024-03328-y
Sparassis latifolia and exercise training as complementary medicine mitigated the 5-fluorouracil potent side effects in mice with colorectal cancer: bioinformatics approaches, novel monitoring pathological metrics, screening signatures, and innovative management tactic
Abstract
Background: Prompt identification and assessment of the disease are essential for reducing the death rate associated with colorectal cancer (COL). Identifying specific causal or sensitive components, such as coding RNA (cRNA) and non-coding RNAs (ncRNAs), may greatly aid in the early detection of colorectal cancer.
Methods: For this purpose, we gave natural chemicals obtained from Sparassis latifolia (SLPs) either alone or in conjunction with chemotherapy (5-Fluorouracil to a mouse colorectal tumor model induced by AOM-DSS. The transcription profile of non-coding RNAs (ncRNAs) and their target hub genes was evaluated using qPCR Real-Time, and ELISA techniques.
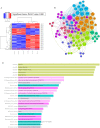
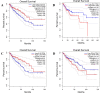
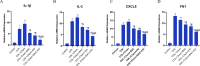
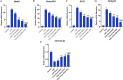
Results: MSX2, MMP7, ITIH4, and COL1A2 were identified as factors in inflammation and oxidative stress, leading to the development of COL. The hub genes listed, upstream regulatory factors such as lncRNA PVT1, NEAT1, KCNQ1OT1, SNHG16, and miR-132-3p have been discovered as biomarkers for prognosis and diagnosis of COL. The SLPs and exercise, effectively decreased the size and quantity of tumors.
Conclusions: This effect may be attributed to the modulation of gene expression levels, including MSX2, MMP7, ITIH4, COL1A2, PVT1, NEAT1, KCNQ1OT1, SNHG16, and miR-132-3p. Ultimately, SLPs and exercise have the capacity to be regarded as complementing and enhancing chemotherapy treatments, owing to their efficacious components.
Keywords: 5-Fluorouracil; AOM; Biomarkers; Colorectal Cancer; DSS; Diagnosis; Exercise; Palliative care; Sparassis latifolia.
© 2024. The Author(s).
Conflict of interest statement
There is no competing of interest to disclose.
Figures









References
LinkOut - more resources
Full Text Sources
Miscellaneous

