The emergence of highly resistant and hypervirulent Klebsiella pneumoniae CC14 clone in a tertiary hospital over 8 years
- PMID: 38637822
- PMCID: PMC11025284
- DOI: 10.1186/s13073-024-01332-5
The emergence of highly resistant and hypervirulent Klebsiella pneumoniae CC14 clone in a tertiary hospital over 8 years
Abstract
Background: Klebsiella pneumoniae is a major bacterial and opportunistic human pathogen, increasingly recognized as a healthcare burden globally. The convergence of resistance and virulence in K. pneumoniae strains has led to the formation of hypervirulent and multidrug-resistant strains with dual risk, limiting treatment options. K. pneumoniae clones are known to emerge locally and spread globally. Therefore, an understanding of the dynamics and evolution of the emerging strains in hospitals is warranted to prevent future outbreaks.
Methods: In this study, we conducted an in-depth genomic analysis on a large-scale collection of 328 multidrug-resistant (MDR) K. pneumoniae strains recovered from 239 patients from a single major hospital in the western coastal city of Jeddah in Saudi Arabia from 2014 through 2022. We employed a broad range of phylogenetic and phylodynamic methods to understand the evolution of the predominant clones on epidemiological time scales, virulence and resistance determinants, and their dynamics. We also integrated the genomic data with detailed electronic health record (EHR) data for the patients to understand the clinical implications of the resistance and virulence of different strains.
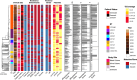
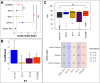
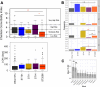
Results: We discovered a diverse population underlying the infections, with most strains belonging to Clonal Complex 14 (CC14) exhibiting dominance. Specifically, we observed the emergence and continuous expansion of strains belonging to the dominant ST2096 in the CC14 clade across hospital wards in recent years. These strains acquired resistance mutations against colistin and extended spectrum β-lactamase (ESBL) and carbapenemase genes, namely blaOXA-48 and blaOXA-232, located on three distinct plasmids, on epidemiological time scales. Strains of ST2096 exhibited a high virulence level with the presence of the siderophore aerobactin (iuc) locus situated on the same mosaic plasmid as the ESBL gene. Integration of ST2096 with EHR data confirmed the significant link between colonization by ST2096 and the diagnosis of sepsis and elevated in-hospital mortality (p-value < 0.05).
Conclusions: Overall, these results demonstrate the clinical significance of ST2096 clones and illustrate the rapid evolution of an emerging hypervirulent and MDR K. pneumoniae in a clinical setting.
Keywords: Klebsiella pneumoniae; Antimicrobial resistance; Precision epidemiology; Whole-genome sequencing.
© 2024. The Author(s).
Conflict of interest statement
None of the authors have any competing interests.
Figures



References
-
- Navon-Venezia S, Kondratyeva K, Carattoli A. Klebsiella pneumoniae: a major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol Rev. 2017;41(3):252–275. - PubMed
-
- GovindarajVaithinathan A, Vanitha A. WHO global priority pathogens list on antibiotic resistance: an urgent need for action to integrate One Health data. Perspect Public Health. 2018;138(2):87–88. - PubMed
-
- Wyres KL, Lam MMC, Holt KE. Population genomics of Klebsiella pneumoniae. Nat Rev Microbiol. 2020;18(6):344–359. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

