Identification of new potent NLRP3 inhibitors by multi-level in-silico approaches
- PMID: 38637900
- PMCID: PMC11027297
- DOI: 10.1186/s13065-024-01178-3
Identification of new potent NLRP3 inhibitors by multi-level in-silico approaches
Abstract
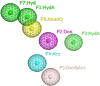
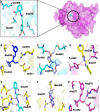
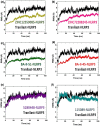
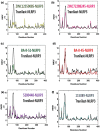
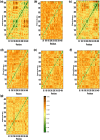
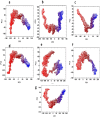
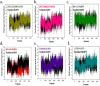
Nod-like receptor protein 3 (NLRP-3), is an intracellular sensor that is involved in inflammasome activation, and the aberrant expression of NLRP3 is responsible for diabetes mellitus, its complications, and many other inflammatory diseases. NLRP3 is considered a promising drug target for novel drug design. Here, a pharmacophore model was generated from the most potent inhibitor, and its validation was performed by the Gunner-Henry scoring method. The validated pharmacophore was used to screen selected compounds databases. As a result, 646 compounds were mapped on the pharmacophore model. After applying Lipinski's rule of five, 391 hits were obtained. All the hits were docked into the binding pocket of target protein. Based on docking scores and interactions with binding site residues, six compounds were selected potential hits. To check the stability of these compounds, 100 ns molecular dynamic (MD) simulations were performed. The RMSD, RMSF, DCCM and hydrogen bond analysis showed that all the six compounds formed stable complex with NLRP3. The binding free energy with the MM-PBSA approach suggested that electrostatic force, and van der Waals interactions, played a significant role in the binding pattern of these compounds. Thus, the outcomes of the current study could provide insights into the identification of new potential NLRP3 inflammasome inhibitors against diabetes and its related disorders.
Keywords: Diabetes mellitus; Inflammasome; Inhibitors; NLRP3; Pharmacophore model.
© 2024. The Author(s).
Conflict of interest statement
The authors have no competing of interests.
Figures
Similar articles
-
Harnessing the Power of Machine Learning Guided Discovery of NLRP3 Inhibitors Towards the Effective Treatment of Rheumatoid Arthritis.Cells. 2024 Dec 30;14(1):27. doi: 10.3390/cells14010027. Cells. 2024. PMID: 39791728 Free PMC article.
-
Pharmacophore modelling, docking and molecular dynamic simulation studies in the discovery of potential human renin inhibitors.J Mol Graph Model. 2022 Nov;116:108272. doi: 10.1016/j.jmgm.2022.108272. Epub 2022 Jul 13. J Mol Graph Model. 2022. PMID: 35932508
-
Identification of novel STAT3 inhibitors for liver fibrosis, using pharmacophore-based virtual screening, molecular docking, and biomolecular dynamics simulations.Sci Rep. 2023 Nov 17;13(1):20147. doi: 10.1038/s41598-023-46193-x. Sci Rep. 2023. PMID: 37978263 Free PMC article.
-
Pharmacophore-Based Virtual Screening and Molecular Dynamics Simulation for Identification of a Novel DNA Gyrase B Inhibitor with Benzoxazine Acetamide Scaffold.ACS Omega. 2021 Dec 22;7(1):1150-1164. doi: 10.1021/acsomega.1c05732. eCollection 2022 Jan 11. ACS Omega. 2021. PMID: 35036778 Free PMC article.
-
Identification of Potent Small-Molecule PCSK9 Inhibitors Based on Quantitative Structure-Activity Relationship, Pharmacophore Modeling, and Molecular Docking Procedure.Curr Probl Cardiol. 2023 Jun;48(6):101660. doi: 10.1016/j.cpcardiol.2023.101660. Epub 2023 Feb 24. Curr Probl Cardiol. 2023. PMID: 36841313 Review.
Cited by
-
High-Throughput Molecular Modeling and Evaluation of the Anti-Inflammatory Potential of Açaí Constituents against NLRP3 Inflammasome.Int J Mol Sci. 2024 Jul 25;25(15):8112. doi: 10.3390/ijms25158112. Int J Mol Sci. 2024. PMID: 39125681 Free PMC article.
-
Phytoconstituents of Chloranthus elatior as a potential adjunct in the treatment of anxiety disorders: In vivo and in silico approaches.Heliyon. 2024 Nov 28;10(23):e40728. doi: 10.1016/j.heliyon.2024.e40728. eCollection 2024 Dec 15. Heliyon. 2024. PMID: 39698094 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
