Diverse potential of secretome from natural killer cells and monocyte-derived macrophages in activating stellate cells
- PMID: 38638443
- PMCID: PMC11025356
- DOI: 10.3389/fimmu.2024.1232070
Diverse potential of secretome from natural killer cells and monocyte-derived macrophages in activating stellate cells
Abstract
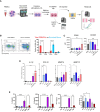
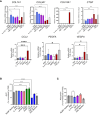
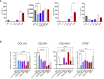
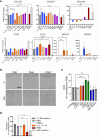
Chronic liver diseases, such as non-alcoholic steatohepatitis (NASH)-induced cirrhosis, are characterized by an increasing accumulation of stressed, damaged, or dying hepatocytes. Hepatocyte damage triggers the activation of resident immune cells, such as Kupffer cells (KC), as well as the recruitment of immune cells from the circulation toward areas of inflammation. After infiltration, monocytes differentiate into monocyte-derived macrophages (MoMF) which are functionally distinct from resident KC. We herein aim to compare the in vitro signatures of polarized macrophages and activated hepatic stellate cells (HSC) with ex vivo-derived disease signatures from human NASH. Furthermore, to shed more light on HSC activation and liver fibrosis progression, we investigate the effects of the secretome from primary human monocytes, macrophages, and NK cells on HSC activation. Interleukin (IL)-4 and IL-13 treatment induced transforming growth factor beta 1 (TGF-β1) secretion by macrophages. However, the supernatant transfer did not induce HSC activation. Interestingly, PMA-activated macrophages showed strong induction of the fibrosis response genes COL10A1 and CTGF, while the supernatant of IL-4/IL-13-treated monocytes induced the upregulation of COL3A1 in HSC. The supernatant of PMA-activated NK cells had the strongest effect on COL10A1 induction in HSC, while IL-15-stimulated NK cells reduced the expression of COL1A1 and CTGF. These data indicate that other factors, aside from the well-known cytokines and chemokines, might potentially be stronger contributors to the activation of HSCs and induction of a fibrotic response, indicating a more diverse and complex role of monocytes, macrophages, and NK cells in liver fibrosis progression.
Keywords: NASH - non-alcoholic steatohepatitis; NK cells; inflammation; liver fibrosis; monocyte-derived macrophage (MDM).
Copyright © 2024 Sauer, Steixner-Kumar, Gabler, Motyka, Rippmann, Brosa, Boettner, Schönberger, Lempp, Frodermann, Simon, Krenkel and Bahrami.
Conflict of interest statement
Author MM is employed by the company Ardigen. Authors JS, AS-K, SG, JR, SB, DB, TS, CL, VF, ES, OK, and EB are employed by the company Boehringer Ingelheim.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

