Mutations in the postsynaptic density signaling hub TNIK disrupt PSD signaling in human models of neurodevelopmental disorders
- PMID: 38638602
- PMCID: PMC11024424
- DOI: 10.3389/fnmol.2024.1359154
Mutations in the postsynaptic density signaling hub TNIK disrupt PSD signaling in human models of neurodevelopmental disorders
Abstract
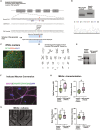
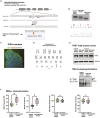
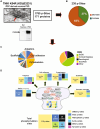
A large number of synaptic proteins have been recurrently associated with complex brain disorders. One of these proteins, the Traf and Nck interacting kinase (TNIK), is a postsynaptic density (PSD) signaling hub, with many variants reported in neurodevelopmental disorder (NDD) and psychiatric disease. While rodent models of TNIK dysfunction have abnormal spontaneous synaptic activity and cognitive impairment, the role of mutations found in patients with TNIK protein deficiency and TNIK protein kinase activity during early stages of neuronal and synapse development has not been characterized. Here, using hiPSC-derived excitatory neurons, we show that TNIK mutations dysregulate neuronal activity in human immature synapses. Moreover, the lack of TNIK protein kinase activity impairs MAPK signaling and protein phosphorylation in structural components of the PSD. We show that the TNIK interactome is enriched in NDD risk factors and TNIK lack of function disrupts signaling networks and protein interactors associated with NDD that only partially overlap to mature mouse synapses, suggesting a differential role of TNIK in immature synapsis in NDD.
Keywords: PSD signaling; glutamatergic neurons; induced pluripotent stem cell (iPSC); neurodevelopment; phosphoproteomics; proteomics.
Copyright © 2024 Jiang, Wilkinson, Flores, Hartel, Mihaylov, Clementel, Flynn, Alkuraya, Ultanir, Graham and Coba.
Conflict of interest statement
The authors declare the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Arons M. H., Thynne C. J., Grabrucker A. M., Li D., Schoen M., Cheyne J. E., et al. . (2012). Autism-associated mutations in ProSAP2/Shank3 impair synaptic transmission and neurexin-neuroligin-mediated transsynaptic signaling. J. Neurosci. 32, 14966–14978. doi: 10.1523/JNEUROSCI.2215-12.2012, PMID: - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

