Integrated bioinformatics analysis reveals upregulated extracellular matrix hub genes in pancreatic cancer: Implications for diagnosis, prognosis, immune infiltration, and therapeutic strategies
- PMID: 38639039
- PMCID: PMC11027013
- DOI: 10.1002/cnr2.2059
Integrated bioinformatics analysis reveals upregulated extracellular matrix hub genes in pancreatic cancer: Implications for diagnosis, prognosis, immune infiltration, and therapeutic strategies
Abstract
Background: Pancreatic cancer (PC) stands out as one of the most formidable malignancies and exhibits an exceptionally unfavorable clinical prognosis due to the absence of well-defined diagnostic indicators and its tendency to develop resistance to therapeutic interventions. The primary objective of this present study was to identify extracellular matrix (ECM)-related hub genes (HGs) and their corresponding molecular signatures, with the intent of potentially utilizing them as biomarkers for diagnostic, prognostic, and therapeutic applications.
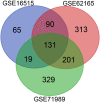
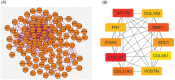
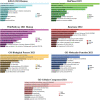
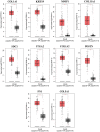
Methods: Three microarray datasets were sourced from the NCBI database to acquire upregulated differentially expressed genes (DEGs), while MatrisomeDB was employed for filtering ECM-related genes. Subsequently, a protein-protein interaction (PPI) network was established using the STRING database. The created network was visually inspected through Cytoscape, and HGs were identified using the CytoHubba plugin tool. Furthermore, enrichment analysis, expression pattern analysis, clinicopathological correlation, survival analysis, immune cell infiltration analysis, and examination of chemical compounds were carried out using Enrichr, GEPIA2, ULCAN, Kaplan Meier plotter, TIMER2.0, and CTD web platforms, respectively. The diagnostic and prognostic significance of HGs was evaluated through the ROC curve analysis.
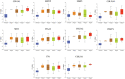
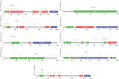
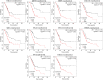
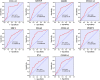
Results: Ten genes associated with ECM functions were identified as HGs among 131 DEGs obtained from microarray datasets. Notably, the expression of these HGs exhibited significantly (p < 0.05) higher in PC, demonstrating a clear association with tumor advancement. Remarkably, higher expression levels of these HGs were inversely correlated with the likelihood of patient survival. Moreover, ROC curve analysis revealed that identified HGs are promising biomarkers for both diagnostic (AUC > 0.75) and prognostic (AUC > 0.64) purposes. Furthermore, we observed a positive correlation between immune cell infiltration and the expression of most HGs. Lastly, our study identified nine compounds with significant interaction profiles that could potentially act as effective chemical agents targeting the identified HGs.
Conclusion: Taken together, our findings suggest that COL1A1, KRT19, MMP1, COL11A1, SDC1, ITGA2, COL1A2, POSTN, FN1, and COL5A1 hold promise as innovative biomarkers for both the diagnosis and prognosis of PC, and they present as prospective targets for therapeutic interventions aimed at impeding the progression PC.
Keywords: collagen proteins; extracellular matrix; hub genes; immune cells infiltration; pancreatic cancer.
© 2024 The Authors. Cancer Reports published by Wiley Periodicals LLC.
Conflict of interest statement
The authors have stated explicitly that there are no conflicts of interest in connection with this article.
Figures




References
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

