M2 Tumor-Associated Macrophages-Derived Exosomal MALAT1 Promotes Glycolysis and Gastric Cancer Progression
- PMID: 38639382
- PMCID: PMC11199979
- DOI: 10.1002/advs.202309298
M2 Tumor-Associated Macrophages-Derived Exosomal MALAT1 Promotes Glycolysis and Gastric Cancer Progression
Abstract
M2-polarized tumor-associated macrophages (M2 TAMs) promote cancer progression. Exosomes mediate cellular communication in the tumor microenvironment (TME). However, the roles of exosomes from M2 TAMs in gastric cancer progression are unclear. Herein, it is reported that M2 TAMs-derived exosomes induced aerobic glycolysis in gastric cancer cells and enhanced their proliferation, metastasis, and chemoresistance in a glycolysis-dependent manner. It is identified that MALAT1 (metastasis-associated lung adenocarcinoma transcript 1) is enriched in M2 TAM exosomes and confirmed that MALAT1 transfer from M2 TAMs to gastric cancer cells via exosomes mediates this effect. Mechanistically, MALAT1 interacted with the δ-catenin protein and suppressed its ubiquitination and degradation by β-TRCP. In addition, MALAT1 upregulated HIF-1α expression by acting as a sponge for miR-217-5p. The activation of β-catenin and HIF-1α signaling pathways by M2 TAM exosomes collectively led to enhanced aerobic glycolysis in gastric cancer cells. Finally, a dual-targeted inhibition of MALAT1 in both gastric cancer cells and macrophages by exosome-mediated delivery of siRNA remarkably suppressed gastric cancer growth and improved chemosensitivity in mouse tumor models. Taken together, these results suggest that M2 TAMs-derived exosomes promote gastric cancer progression via MALAT1-mediated regulation of glycolysis. The findings offer a potential target for gastric cancer therapy.
Keywords: MALAT1; exosomes; gastric cancer; glycolysis; tumor‐associated macrophages.
© 2024 The Authors. Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


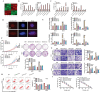
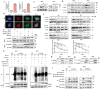




References
MeSH terms
Substances
Grants and funding
- 82372909/National Natural Science Foundation of China
- 81972310/National Natural Science Foundation of China
- BK20200043/Distinguished Young Scholar Project of Jiangsu Province
- 2019GSZDSYS01/Key Laboratory of Molecular Diagnostics and Precision Medicine for Surgical Oncology in Gansu Province
- 2019GSZDSYS02/Key Laboratory of Molecular Diagnostics and Precision Medicine for Surgical Oncology in Gansu Province
LinkOut - more resources
Full Text Sources
Medical
