A modified BCG with depletion of enzymes associated with peptidoglycan amidation induces enhanced protection against tuberculosis in mice
- PMID: 38639995
- PMCID: PMC11132681
- DOI: 10.7554/eLife.89157
A modified BCG with depletion of enzymes associated with peptidoglycan amidation induces enhanced protection against tuberculosis in mice
Abstract
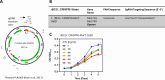
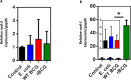
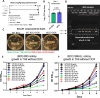
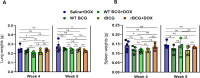
Mechanisms by which Mycobacterium tuberculosis (Mtb) evades pathogen recognition receptor activation during infection may offer insights for the development of improved tuberculosis (TB) vaccines. Whilst Mtb elicits NOD-2 activation through host recognition of its peptidoglycan-derived muramyl dipeptide (MDP), it masks the endogenous NOD-1 ligand through amidation of glutamate at the second position in peptidoglycan side-chains. As the current BCG vaccine is derived from pathogenic mycobacteria, a similar situation prevails. To alleviate this masking ability and to potentially improve efficacy of the BCG vaccine, we used CRISPRi to inhibit expression of the essential enzyme pair, MurT-GatD, implicated in amidation of peptidoglycan side-chains. We demonstrate that depletion of these enzymes results in reduced growth, cell wall defects, increased susceptibility to antibiotics, altered spatial localization of new peptidoglycan and increased NOD-1 expression in macrophages. In cell culture experiments, training of a human monocyte cell line with this recombinant BCG yielded improved control of Mtb growth. In the murine model of TB infection, we demonstrate that depletion of MurT-GatD in BCG, which is expected to unmask the D-glutamate diaminopimelate (iE-DAP) NOD-1 ligand, yields superior prevention of TB disease compared to the standard BCG vaccine. In vitro and in vivo experiments in this study demonstrate the feasibility of gene regulation platforms such as CRISPRi to alter antigen presentation in BCG in a bespoke manner that tunes immunity towards more effective protection against TB disease.
Keywords: NOD1; amidation; infectious disease; microbiology; mouse; mycobacterium; peptidoglycan; recombinant BCG vaccine; tuberculosis.
Plain language summary
Tuberculosis is the leading cause of death from an infectious disease worldwide, partially due to a lack of access to drug treatments in certain countries where the disease is common. The only available tuberculosis vaccine – known as the BCG vaccine – is useful for preventing cases in young children, but is ineffective in teenagers and adults. So, there is a need to develop new vaccines that offer better, and longer lasting, durable protection in people of all ages. During an infection, our immune system recognizes markers known as PAMPs on the surface of bacteria, viruses or other disease-causing pathogens. The recognition of PAMPs by the immune system enables the body to distinguish foreign invading organisms from its own cells and tissues, thus triggering a response that fights the infection. If the body encounters the infectious agent again in the future, the immune system is able to quickly recognize and eliminate it before it can cause disease. Vaccines protect us by mimicking the appearance of the pathogen to trigger the first immune response without causing the illness. The BCG vaccine contains live bacteria that are closely related to the bacterium responsible for tuberculosis called Mycobacterium tuberculosis. Both M. tuberculosis and the live bacteria used in the BCG vaccine are able to hide an important PAMP, known as the NOD-1 ligand, from the immune system, making it harder for the body to detect them. The NOD-1 ligand forms part of the bacterial cell wall and modifying the BCG bacterium so it cannot disguise this PAMP may lead to a new, more effective vaccine. To investigate this possibility, Shaku et al. used a gene editing approach to develop a modified version of the BCG bacterium which is unable to hide its NOD-1 ligand when treated with a specific drug. Immune cells trained with the modified BCG vaccine were more effective at controlling the growth of M. tuberculosis than macrophages trained using the original vaccine. Furthermore, mice vaccinated with the modified BCG vaccine were better able to limit M. tuberculosis growth in their lungs than mice that had received the original vaccine. These findings offer a new candidate vaccine in the fight against tuberculosis. Further studies will be needed to modify the vaccine for use in humans. More broadly, this work demonstrates that gene editing can be used to expose a specific PAMP present in a live vaccine. This may help develop more effective vaccines for other diseases in the future.
© 2024, Shaku et al.
Conflict of interest statement
MS, PU, KO, AA, MP, WB No competing interests declared, BK Senior editor, eLife
Figures









Update of
-
A modified BCG with depletion of enzymes associated with peptidoglycan amidation induces enhanced protection against tuberculosis in mice.bioRxiv [Preprint]. 2023 May 3:2023.05.03.539199. doi: 10.1101/2023.05.03.539199. bioRxiv. 2023. Update in: Elife. 2024 Apr 19;13:e89157. doi: 10.7554/eLife.89157. PMID: 37205421 Free PMC article. Updated. Preprint.
References
-
- Angelidou A, Diray-Arce J, Conti MG, Smolen KK, van Haren SD, Dowling DJ, Husson RN, Levy O. BCG as a case study for precision vaccine development: lessons from vaccine heterogeneity, trained immunity, and immune ontogeny. Frontiers in Microbiology. 2020;11:332. doi: 10.3389/fmicb.2020.00332. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

