Decoding the role of oxidative stress resistance and alternative carbon substrate assimilation in the mature biofilm growth mode of Candida glabrata
- PMID: 38641593
- PMCID: PMC11031924
- DOI: 10.1186/s12866-024-03274-9
Decoding the role of oxidative stress resistance and alternative carbon substrate assimilation in the mature biofilm growth mode of Candida glabrata
Abstract
Background: Biofilm formation is viewed as a vital mechanism in C. glabrata pathogenesis. Although, it plays a significant role in virulence but transcriptomic architecture and metabolic pathways governing the biofilm growth mode of C. glabrata remain elusive. The present study intended to investigate the genes implicated in biofilm growth phase of C. glabrata through global transcriptomic approach.
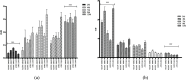
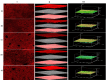
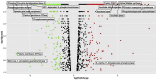
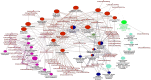
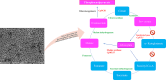
Results: Functional analysis of Differentially expressed genes (DEGs) using gene ontology and pathways analysis revealed that upregulated genes are involved in the glyoxylate cycle, carbon-carbon lyase activity, pre-autophagosomal structure membrane and vacuolar parts whereas, down- regulated genes appear to be associated with glycolysis, ribonucleoside biosynthetic process, ribosomal and translation process in the biofilm growth condition. The RNA-Seq expression of eight selected DEGs (CgICL1, CgMLS1, CgPEP1, and CgNTH1, CgERG9, CgERG11, CgTEF3, and CgCOF1) was performed with quantitative real-time PCR (RT-qPCR). The gene expression profile of selected DEGs with RT-qPCR displayed a similar pattern of expression as observed in RNA-Seq. Phenotype screening of mutant strains generated for genes CgPCK1 and CgPEP1, showed that Cgpck1∆ failed to grow on alternative carbon substrate (Glycerol, Ethanol, Oleic acid) and similarly, Cgpep1∆ unable to grow on YPD medium supplemented with hydrogen peroxide. Our results suggest that in the absence of glucose, C. glabrata assimilate glycerol, oleic acid and generate acetyl coenzyme-A (acetyl-CoA) which is a central and connecting metabolite between catabolic and anabolic pathways (glyoxylate and gluconeogenesis) to produce glucose and fulfil energy requirements.
Conclusions: The study was executed using various approaches (transcriptomics, functional genomics and gene deletion) and it revealed that metabolic plasticity of C. glabrata (NCCPF-100,037) in biofilm stage modulates its virulence and survival ability to counter the stress and may promote its transition from commensal to opportunistic pathogen. The observations deduced from the present study along with future work on characterization of the proteins involved in this intricate process may prove to be beneficial for designing novel antifungal strategies.
Keywords: C. glabrata; Alternative carbon substrate assimilation; Biofilm; Oxidative stress; Transcriptomics.
© 2024. The Author(s).
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

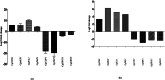


References
-
- Pfaller MA, Andes DR, Diekema DJ, Horn DL, Reboli AC, Rotstein C, et al. Epidemiology and outcomes of invasive candidiasis due to non-albicans species of Candida in 2,496 patients: data from the prospective antifungal therapy (PATH) registry 2004–2008. PLoS ONE. 2014;9:e101510. doi: 10.1371/journal.pone.0101510. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

