Revealing Prdx4 as a potential diagnostic and therapeutic target for acute pancreatitis based on machine learning analysis
- PMID: 38641608
- PMCID: PMC11027343
- DOI: 10.1186/s12920-024-01854-2
Revealing Prdx4 as a potential diagnostic and therapeutic target for acute pancreatitis based on machine learning analysis
Abstract
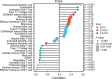
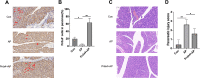
Acute pancreatitis (AP) is a common systemic inflammatory disease resulting from the activation of trypsinogen by various incentives in ICU. The annual incidence rate is approximately 30 out of 100,000. Some patients may progress to severe acute pancreatitis, with a mortality rate of up to 40%. Therefore, the goal of this article is to explore the key genes for effective diagnosis and treatment of AP. The analysis data for this study were merged from two GEO datasets. 1357 DEGs were used for functional enrichment and cMAP analysis, aiming to reveal the pathogenic genes and potential mechanisms of AP, as well as potential drugs for treating AP. Importantly, the study used LASSO and SVM-RFE machine learning to screen the most likely AP occurrence biomarker for Prdx4 among numerous candidate genes. A receiver operating characteristic of Prdx4 was used to estimate the incidence of AP. The ssGSEA algorithm was employed to investigate immune cell infiltration in AP. The biomarker Prdx4 gene exhibited significant associations with a majority of immune cells and was identified as being expressed in NKT cells, macrophages, granulocytes, and B cells based on single-cell transcriptome data. Finally, we found an increase in Prdx4 expression in the pancreatic tissue of AP mice through immunohistochemistry. After treatment with recombinant Prdx4, the pathological damage to the pancreatic tissue of AP mice was relieved. In conclusion, our study identified Prdx4 as a potential AP hub gene, providing a new target for treatment.
Keywords: Acute pancreatitis (AP); Bioinformatics analysis; Diagnostic value; Immune cell infiltration; Machine learning.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures







Similar articles
-
Investigating potential biomarkers of acute pancreatitis in patients with a BMI>30 using Mendelian randomization and transcriptomic analysis.Lipids Health Dis. 2024 Apr 22;23(1):119. doi: 10.1186/s12944-024-02102-3. Lipids Health Dis. 2024. PMID: 38649912 Free PMC article.
-
Exploring the pathogenesis, biomarkers, and potential drugs for type 2 diabetes mellitus and acute pancreatitis through a comprehensive bioinformatic analysis.Front Endocrinol (Lausanne). 2024 Nov 20;15:1405726. doi: 10.3389/fendo.2024.1405726. eCollection 2024. Front Endocrinol (Lausanne). 2024. PMID: 39634181 Free PMC article.
-
Identification and validation of novel biomarkers associated with immune infiltration for the diagnosis of osteosarcoma based on machine learning.Front Genet. 2023 Sep 4;14:1136783. doi: 10.3389/fgene.2023.1136783. eCollection 2023. Front Genet. 2023. PMID: 37732314 Free PMC article.
-
Screening and identification of the hub genes in severe acute pancreatitis and sepsis.Front Mol Biosci. 2024 Sep 19;11:1425143. doi: 10.3389/fmolb.2024.1425143. eCollection 2024. Front Mol Biosci. 2024. PMID: 39364223 Free PMC article.
-
Identification of novel biomarkers and immune infiltration characteristics of ischemic stroke based on comprehensive bioinformatic analysis and machine learning.Biochem Biophys Rep. 2023 Dec 7;37:101595. doi: 10.1016/j.bbrep.2023.101595. eCollection 2024 Mar. Biochem Biophys Rep. 2023. PMID: 38371524 Free PMC article.
References
-
- Bourgault J, Abner E, Manikpurage HD, Pujol-Gualdo N, Laisk T, Gobeil É, Gagnon E, Girard A, Mitchell PL, Thériault S, et al. Proteome-wide mendelian randomization identifies causal links between blood proteins and Acute Pancreatitis. Gastroenterology. 2023;164(6):953–e965953. doi: 10.1053/j.gastro.2023.01.028. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 2021LCYB12/Clinical Research cultivation Program of the Second Affiliated Hospital of Anhui Medical University
- gxyq2022006/Support Program for Elite Young Talents in Colleges and Universities of Anhui Province
- 2022GMFY10/National Natural Science Foundation Incubation Program of The Second Affiliated Hospital of Anhui Medical University
- 2022GMFY09/National Natural Science Foundation Incubation Program of The Second Affiliated Hospital of Anhui Medical University
- 2022jyxm750/the Provincial Quality Engineering Project of Higher Education Institutions of Anhui Province
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

