Hepatocytes coordinate immune evasion in cancer via release of serum amyloid A proteins
- PMID: 38641718
- PMCID: PMC11186515
- DOI: 10.1038/s41590-024-01820-1
Hepatocytes coordinate immune evasion in cancer via release of serum amyloid A proteins
Abstract
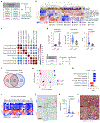
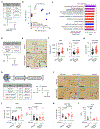
T cell infiltration into tumors is a favorable prognostic feature, but most solid tumors lack productive T cell responses. Mechanisms that coordinate T cell exclusion are incompletely understood. Here we identify hepatocyte activation via interleukin-6/STAT3 and secretion of serum amyloid A (SAA) proteins 1 and 2 as important regulators of T cell surveillance of extrahepatic tumors. Loss of STAT3 in hepatocytes or SAA remodeled the tumor microenvironment with infiltration by CD8+ T cells, while interleukin-6 overexpression in hepatocytes and SAA signaling via Toll-like receptor 2 reduced the number of intratumoral dendritic cells and, in doing so, inhibited T cell tumor infiltration. Genetic ablation of SAA enhanced survival after tumor resection in a T cell-dependent manner. Likewise, in individuals with pancreatic ductal adenocarcinoma, long-term survivors after surgery demonstrated lower serum SAA levels than short-term survivors. Taken together, these data define a fundamental link between liver and tumor immunobiology wherein hepatocytes govern productive T cell surveillance in cancer.
© 2024. The Author(s), under exclusive licence to Springer Nature America, Inc.
Figures












References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

