Computational analysis of affinity dynamics between the variants of SARS-CoV-2 spike protein (RBD) and human ACE-2 receptor
- PMID: 38641844
- PMCID: PMC11031966
- DOI: 10.1186/s12985-024-02365-3
Computational analysis of affinity dynamics between the variants of SARS-CoV-2 spike protein (RBD) and human ACE-2 receptor
Abstract
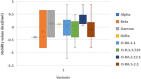
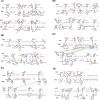
The novel coronavirus SARS-CoV-2 resulted in a significant worldwide health emergency known as the COVID-19 pandemic. This crisis has been marked by the widespread of various variants, with certain ones causing notable apprehension. In this study, we harnessed computational techniques to scrutinize these Variants of Concern (VOCs), including various Omicron subvariants. Our approach involved the use of protein structure prediction algorithms and molecular docking techniques, we have investigated the effects of mutations within the Receptor Binding Domain (RBD) of SARS-CoV-2 and how these mutations influence its interactions with the human angiotensin-converting enzyme 2 (hACE-2) receptor. Further we have predicted the structural alterations in the RBD of naturally occurring SARS-CoV-2 variants using the tr-Rosetta algorithm. Subsequent docking and binding analysis employing HADDOCK and PRODIGY illuminated crucial interactions occurring at the Receptor-Binding Motif (RBM). Our findings revealed a hierarchy of increased binding affinity between the human ACE2 receptor and the various RBDs, in the order of wild type (Wuhan-strain) < Beta < Alpha < Gamma < Omicron-B.1.1.529 < Delta < Omicron-BA.2.12.1 < Omicron-BA.5.2.1 < Omicron-BA.1.1. Notably, Omicron-BA.1.1 demonstrated the highest binding affinity of -17.4 kcal mol-1 to the hACE2 receptor when compared to all the mutant complexes. Additionally, our examination indicated that mutations occurring in active residues of the Receptor Binding Domain (RBD) consistently improved the binding affinity and intermolecular interactions in all mutant complexes. Analysis of the differences among variants has laid a foundation for the structure-based drug design targeting the RBD region of SARS-CoV-2.
Keywords: HADDOCK; SARS-CoV-2; Variants of concern (VOCs); h-ACE2; trRosetta.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
In silico study of SARS-CoV-2 spike protein RBD and human ACE-2 affinity dynamics across variants and Omicron subvariants.J Med Virol. 2023 Jan;95(1):e28406. doi: 10.1002/jmv.28406. J Med Virol. 2023. PMID: 36519577 Free PMC article.
-
Structural basis and analysis of hamster ACE2 binding to different SARS-CoV-2 spike RBDs.J Virol. 2024 Mar 19;98(3):e0115723. doi: 10.1128/jvi.01157-23. Epub 2024 Feb 2. J Virol. 2024. PMID: 38305152 Free PMC article.
-
Computational modeling of the effect of five mutations on the structure of the ACE2 receptor and their correlation with infectivity and virulence of some emerged variants of SARS-CoV-2 suggests mechanisms of binding affinity dysregulation.Chem Biol Interact. 2022 Dec 1;368:110244. doi: 10.1016/j.cbi.2022.110244. Epub 2022 Nov 3. Chem Biol Interact. 2022. PMID: 36336003 Free PMC article.
-
Surface charge changes in spike RBD mutations of SARS-CoV-2 and its variant strains alter the virus evasiveness via HSPGs: A review and mechanistic hypothesis.Front Public Health. 2022 Aug 24;10:952916. doi: 10.3389/fpubh.2022.952916. eCollection 2022. Front Public Health. 2022. PMID: 36091499 Free PMC article. Review.
-
Unique mutations in SARS-CoV-2 Omicron subvariants' non-spike proteins: Potential impacts on viral pathogenesis and host immune evasion.Microb Pathog. 2022 Sep;170:105699. doi: 10.1016/j.micpath.2022.105699. Epub 2022 Aug 6. Microb Pathog. 2022. PMID: 35944840 Free PMC article. Review.
Cited by
-
Uncovering SARS-CoV-2 Molecular Epidemiology Across the Pandemic Transition: Insights into Transmission in Clinical and Environmental Samples.Viruses. 2025 May 19;17(5):726. doi: 10.3390/v17050726. Viruses. 2025. PMID: 40431737 Free PMC article.
References
-
- Chu Y-M, Zarin R, Khan A, Murtaza S. A vigorous study of fractional order mathematical model for SARS-CoV-2 epidemic with Mittag-Leffler kernel. Alexandria Eng J. 2023;71:565–79. doi: 10.1016/j.aej.2023.03.037. - DOI
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

