spVC for the detection and interpretation of spatial gene expression variation
- PMID: 38641849
- PMCID: PMC11027374
- DOI: 10.1186/s13059-024-03245-3
spVC for the detection and interpretation of spatial gene expression variation
Abstract
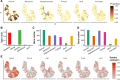
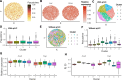
Spatially resolved transcriptomics technologies have opened new avenues for understanding gene expression heterogeneity in spatial contexts. However, existing methods for identifying spatially variable genes often focus solely on statistical significance, limiting their ability to capture continuous expression patterns and integrate spot-level covariates. To address these challenges, we introduce spVC, a statistical method based on a generalized Poisson model. spVC seamlessly integrates constant and spatially varying effects of covariates, facilitating comprehensive exploration of gene expression variability and enhancing interpretability. Simulation and real data applications confirm spVC's accuracy in these tasks, highlighting its versatility in spatial transcriptomics analysis.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
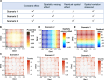

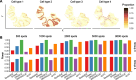
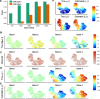
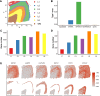


Comment on
-
Spatial Transcriptomics and In Situ Sequencing to Study Alzheimer's Disease.Cell. 2020 Aug 20;182(4):976-991.e19. doi: 10.1016/j.cell.2020.06.038. Epub 2020 Jul 22. Cell. 2020. PMID: 32702314

