Myosin inhibitor reverses hypertrophic cardiomyopathy in genotypically diverse pediatric iPSC-cardiomyocytes to mirror variant correction
- PMID: 38642550
- PMCID: PMC11148569
- DOI: 10.1016/j.xcrm.2024.101520
Myosin inhibitor reverses hypertrophic cardiomyopathy in genotypically diverse pediatric iPSC-cardiomyocytes to mirror variant correction
Abstract
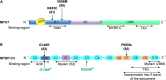
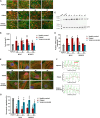
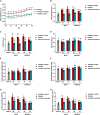
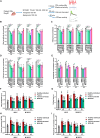
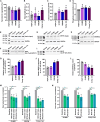
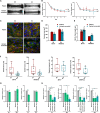
Pathogenic variants in MYH7 and MYBPC3 account for the majority of hypertrophic cardiomyopathy (HCM). Targeted drugs like myosin ATPase inhibitors have not been evaluated in children. We generate patient and variant-corrected iPSC-cardiomyocytes (CMs) from pediatric HCM patients harboring single variants in MYH7 (V606M; R453C), MYBPC3 (G148R) or digenic variants (MYBPC3 P955fs, TNNI3 A157V). We also generate CMs harboring MYBPC3 mono- and biallelic variants using CRISPR editing of a healthy control. Compared with isogenic and healthy controls, variant-positive CMs show sarcomere disorganization, higher contractility, calcium transients, and ATPase activity. However, only MYH7 and biallelic MYBPC3 variant-positive CMs show stronger myosin-actin binding. Targeted myosin ATPase inhibitors show complete rescue of the phenotype in variant-positive CMs and in cardiac Biowires to mirror isogenic controls. The response is superior to verapamil or metoprolol. Myosin inhibitors can be effective in genotypically diverse HCM highlighting the need for myosin inhibitor drug trials in pediatric HCM.
Keywords: CRISPR gene editing; MYBPC3; MYH7; cardiomyocytes; hypertrophic cardiomyopathy; induced pluripotent stem cells; metoprolol; myosin ATPase inhibitor; pediatric; verapamil.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests S.M. is a consultant for Bristol Myers Squibb and Tenaya Therapeutics. M.R. and Y.Z. are inventors on an issued US patent covering Biowire tissue fabrication. They receive royalties from Valo Health. M.R. has a consulting agreement with Valo Health and had a consulting agreement with Tenaya Therapeutics. M.R. and Y.Z. are co-founders of TARA Biosystems Inc. and held equity in the company until April 2022.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

