High LGALS3 expression induced by HCP5/hsa-miR-27b-3p correlates with poor prognosis and tumor immune infiltration in hepatocellular carcinoma
- PMID: 38643145
- PMCID: PMC11031979
- DOI: 10.1186/s12935-024-03309-1
High LGALS3 expression induced by HCP5/hsa-miR-27b-3p correlates with poor prognosis and tumor immune infiltration in hepatocellular carcinoma
Abstract
Background: Hepatocellular carcinoma (HCC) is widely recognized for its unfavorable prognosis. Increasing evidence has revealed that LGALS3 has an essential function in initiating and developing several malignancies in humans. Nevertheless, thorough analysis of the expression profile, clinical prognosis, pathway prediction, and immune infiltration of LGALS3 has not been fully explored in HCC.
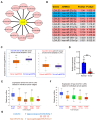
Methods: In this study, an initial pan-cancer analysis was conducted to investigate the expression and prognosis of LGALS3. Following a comprehensive analysis, which included expression analysis and correlation analysis, noncoding RNAs that contribute to the overexpression of LGALS3 were subsequently identified. This identification was further validated using HCC clinical tissue samples. TIMER2 and GEPIA2 were employed to examine the correlation between LGALS3 and HCP5 with immunological checkpoints, cell chemotaxis, and immune infiltration in HCC. The R program was applied to analyze the expression distribution of immune score in in HCC patients with high and low LGALS3 expression. The expression profiles of immune checkpoints were also analyzed. Use R to perform GSVA analysis in order to explore potential signaling pathways.
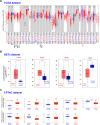
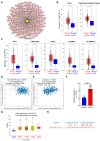
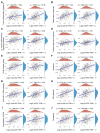
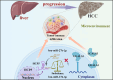
Results: First, we conducted pan-cancer analysis for LGALS3 expression level through an in-depth analysis of public databases and found that HCC has a high LGALS3 gene and protein expression level, which were then verified in clinical HCC specimens. Meanwhile, high LGALS3 gene expression is related to malignant progression and poor prognosis of HCC. Univariate and multivariate analyses confirmed that LGALS3 could serve as an independent prognostic marker for HCC. Next, by combining comprehensive analysis and validation on HCC clinical tissue samples, we hypothesize that the HCP5/hsa-miR-27b-3p axis could serve as the most promising LGALS3 regulation mechanism in HCC. KEGG and GO analyses highlighted that the LGALS3-related genes were involved in tumor immunity. Furthermore, we detected a significant positive association between LGALS3 and HCP5 with immunological checkpoints, cell chemotaxis, and immune infiltration. In addition, high LGALS3 expression groups had significantly higher immune cell scores and immune checkpoint expression levels. Finally, GSVA analysis was performed to predict potential signaling pathways linked to LGALS3 and HCP5 in immune evasion and metabolic reprogramming of HCC.
Conclusions: Our findings indicated that the upregulation of LGALS3 via the HCP5/hsa-miR-27b-3p axis is associated with unfavorable prognosis and increased tumor immune infiltration in HCC.
Keywords: HCP5; Hepatocellular carcinoma; Immune cell infiltration; LGALS3; Prognosis; Tumor microenvironment.
© 2024. The Author(s).
Conflict of interest statement
The authors state no competing interests.
Figures




References
-
- Khan TH, Muhammad N, Tarique M, Usmani D, Naz H, Sarode A. The role of Cancer-Specific Target antigens in CAR T Cell Therapy in Hematological Malignancies. Current Tissue Microenvironment Reports; 2024.
Grants and funding
- 21JCQNJC00150/Natural Science Foundation of Tianjin
- TJWJ2022QN026/Science and Technology Foundation of Tianjin Health Bureau
- 22JCZDJC00450/Key Project of Natural Science Foundation of Tianjin
- TJWJ2021MS006/Project of Health Commission of Tianjin
- TJYXZDXK-045A/Tianjin Key Medical Discipline (Specialty) Construction Project
LinkOut - more resources
Full Text Sources

