Metabolic and inflammatory perturbation of diabetes associated gut dysbiosis in people living with and without HIV infection
- PMID: 38643166
- PMCID: PMC11032597
- DOI: 10.1186/s13073-024-01336-1
Metabolic and inflammatory perturbation of diabetes associated gut dysbiosis in people living with and without HIV infection
Abstract
Background: Gut dysbiosis has been linked with both HIV infection and diabetes, but its interplay with metabolic and inflammatory responses in diabetes, particularly in the context of HIV infection, remains unclear.
Methods: We first conducted a cross-sectional association analysis to characterize the gut microbial, circulating metabolite, and immune/inflammatory protein features associated with diabetes in up to 493 women (~ 146 with prevalent diabetes with 69.9% HIV +) of the Women's Interagency HIV Study. Prospective analyses were then conducted to determine associations of identified metabolites with incident diabetes over 12 years of follow-up in 694 participants (391 women from WIHS and 303 men from the Multicenter AIDS Cohort Study; 166 incident cases were recorded) with and without HIV infection. Mediation analyses were conducted to explore whether gut bacteria-diabetes associations are explained by altered metabolites and proteins.
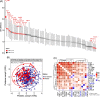
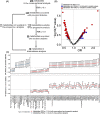
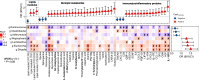
Results: Seven gut bacterial genera were identified to be associated with diabetes (FDR-q < 0.1), with positive associations for Shigella, Escherichia, Megasphaera, and Lactobacillus, and inverse associations for Adlercreutzia, Ruminococcus, and Intestinibacter. Importantly, the associations of most species, especially Adlercreutzia and Ruminococcus, were largely independent of antidiabetic medications use. Meanwhile, 18 proteins and 76 metabolites, including 3 microbially derived metabolites (trimethylamine N-oxide, phenylacetylglutamine (PAGln), imidazolepropionic acid (IMP)), 50 lipids (e.g., diradylglycerols (DGs) and triradylglycerols (TGs)) and 23 non-lipid metabolites, were associated with diabetes (FDR-q < 0.1), with the majority showing positive associations and more than half of them (59/76) associated with incident diabetes. In mediation analyses, several proteins, especially interleukin-18 receptor 1 and osteoprotegerin, IMP and PAGln partially mediate the observed bacterial genera-diabetes associations, particularly for those of Adlercreutzia and Escherichia. Many diabetes-associated metabolites and proteins were altered in HIV, but no effect modification on their associations with diabetes was observed by HIV.
Conclusion: Among individuals with and without HIV, multiple gut bacterial genera, blood metabolites, and proinflammatory proteins were associated with diabetes. The observed mediated effects by metabolites and proteins in genera-diabetes associations highlighted the potential involvement of inflammatory and metabolic perturbations in the link between gut dysbiosis and diabetes in the context of HIV infection.
Keywords: Blood metabolome; Diabetes; Gut dysbiosis; Gut metagenome; HIV infection; Inflammatory proteome; Multi-omics integration.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential competing interest.
Figures



References
-
- Aron-Wisnewsky J, Warmbrunn MV, Nieuwdorp M, Clement K. Metabolism and mMetabolic dDisorders and the mMicrobiome: the intestinal microbiota associated with obesity, lipid metabolism, and metabolic health-pathophysiology and therapeutic strategies. Gastroenterology. 2021;160:573–599. doi: 10.1053/j.gastro.2020.10.057. - DOI - PubMed
Publication types
MeSH terms
Grants and funding
- P30 DK040561/DK/NIDDK NIH HHS/United States
- R01HL140976/HL/NHLBI NIH HHS/United States
- U01 AI035004/AI/NIAID NIH HHS/United States
- U01 HL146208/HL/NHLBI NIH HHS/United States
- R01 HL126543/HL/NHLBI NIH HHS/United States
- U01 HL146192/HL/NHLBI NIH HHS/United States
- R01DK119268/DK/NIDDK NIH HHS/United States
- P30 AI124414/AI/NIAID NIH HHS/United States
- U01 HL146194/HL/NHLBI NIH HHS/United States
- U01 HL146241/HL/NHLBI NIH HHS/United States
- P30 AI027767/AI/NIAID NIH HHS/United States
- P30 AI050409/AI/NIAID NIH HHS/United States
- U01 HL146333/HL/NHLBI NIH HHS/United States
- R01 DK126698/DK/NIDDK NIH HHS/United States
- U01 HL146245/HL/NHLBI NIH HHS/United States
- R01 MD011389/MD/NIMHD NIH HHS/United States
- R01 DK119268/DK/NIDDK NIH HHS/United States
- U01 HL146205/HL/NHLBI NIH HHS/United States
- R01 HL095140/HL/NHLBI NIH HHS/United States
- P30 MH116867/MH/NIMH NIH HHS/United States
- R01 HL083760/HL/NHLBI NIH HHS/United States
- R01 HL132794/HL/NHLBI NIH HHS/United States
- K01 HL169019/HL/NHLBI NIH HHS/United States
- U01 HL146242/HL/NHLBI NIH HHS/United States
- R01 HL170904/HL/NHLBI NIH HHS/United States
- K01 HL129892/HL/NHLBI NIH HHS/United States
- P30 DK111022/DK/NIDDK NIH HHS/United States
- P30 AI073961/AI/NIAID NIH HHS/United States
- U01 HL146201/HL/NHLBI NIH HHS/United States
- U01 HL146193/HL/NHLBI NIH HHS/United States
- R01 HL140976/HL/NHLBI NIH HHS/United States
- U01 HL146204/HL/NHLBI NIH HHS/United States
- U01 HL146202/HL/NHLBI NIH HHS/United States
- UL1 TR001881/TR/NCATS NIH HHS/United States
- P30 CA013330/CA/NCI NIH HHS/United States
- UL1 TR000004/TR/NCATS NIH HHS/United States
- U01 HL146240/HL/NHLBI NIH HHS/United States
- U01 HL146203/HL/NHLBI NIH HHS/United States
- K01 HL137557/HL/NHLBI NIH HHS/United States
- UL1 TR003098/TR/NCATS NIH HHS/United States
- P30 AI050410/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical

