High glucose-induced p66Shc mitochondrial translocation regulates autophagy initiation and autophagosome formation in syncytiotrophoblast and extravillous trophoblast
- PMID: 38643181
- PMCID: PMC11031965
- DOI: 10.1186/s12964-024-01621-x
High glucose-induced p66Shc mitochondrial translocation regulates autophagy initiation and autophagosome formation in syncytiotrophoblast and extravillous trophoblast
Abstract
Background: p66Shc, as a redox enzyme, regulates reactive oxygen species (ROS) production in mitochondria and autophagy. However, the mechanisms by which p66Shc affects autophagosome formation are not fully understood.
Methods: p66Shc expression and its location in the trophoblast cells were detected in vivo and in vitro. Small hairpin RNAs or CRISPR/Cas9, RNA sequencing, and confocal laser scanning microscope were used to clarify p66Shc's role in regulating autophagic flux and STING activation. In addition, p66Shc affects mitochondrial-associated endoplasmic reticulum membranes (MAMs) formation were observed by transmission electron microscopy (TEM). Mitochondrial function was evaluated by detected cytoplastic mitochondrial DNA (mtDNA) and mitochondrial membrane potential (MMP).
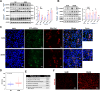
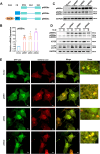
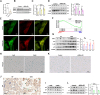
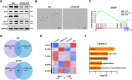
Results: High glucose induces the expression and mitochondrial translocation of p66Shc, which promotes MAMs formation and stimulates PINK1-PRKN-mediated mitophagy. Moreover, mitochondrial localized p66Shc reduces MMP and triggers cytosolic mtDNA release, thus activates cGAS/STING signaling and ultimately leads to enhanced autophagy and cellular senescence. Specially, we found p66Shc is required for the interaction between STING and LC3II, as well as between STING and ATG5, thereby regulates cGAS/STING-mediated autophagy. We also identified hundreds of genes associated several biological processes including aging are co-regulated by p66Shc and ATG5, deletion either of which results in diminished cellular senescence.
Conclusion: p66Shc is not only implicated in the initiation of autophagy by promoting MAMs formation, but also helps stabilizing active autophagic flux by activating cGAS/STING pathway in trophoblast.
Keywords: Autophagy; High glucose; MAM; cGAS/STING; p66Shc.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Nakashima A, Yamanaka-Tatematsu M, Fujita N, Koizumi K, Shima T, Yoshida T, Nikaido T, Okamoto A, Yoshimori T, Saito S. Impaired autophagy by soluble endoglin, under physiological hypoxia in early pregnant period, is involved in poor placentation in preeclampsia. Autophagy. 2013;9:303–316. doi: 10.4161/auto.22927. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

