Genetic Tools for Cell Lineage Tracing and Profiling Developmental Trajectories in the Skin
- PMID: 38643988
- PMCID: PMC11034889
- DOI: 10.1016/j.jid.2024.02.006
Genetic Tools for Cell Lineage Tracing and Profiling Developmental Trajectories in the Skin
Abstract
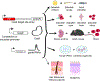
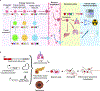
The epidermis is the body's first line of protection against dehydration and pathogens, continually regenerating the outermost protective skin layers throughout life. During both embryonic development and wound healing, epidermal stem and progenitor cells must respond to external stimuli and insults to build, maintain, and repair the cutaneous barrier. Recent advances in CRISPR-based methods for cell lineage tracing have remarkably expanded the potential for experiments that track stem and progenitor cell proliferation and differentiation over the course of tissue and even organismal development. Additional tools for DNA-based recording of cellular signaling cues promise to deepen our understanding of the mechanisms driving normal skin morphogenesis and response to stressors as well as the dysregulation of cell proliferation and differentiation in skin diseases and cancer. In this review, we highlight cutting-edge methods for cell lineage tracing, including in organoids and model organisms, and explore how cutaneous biology researchers might leverage these techniques to elucidate the developmental programs that support the regenerative capacity and plasticity of the skin.
Keywords: CRISPR; DNA barcoding; DNA typewriter; Genome editing; Prime editor.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
CONFLICT OF INTEREST
The University of Washington has filed a patent application for several technologies related to molecular recording, including DNA Typewriter, ENGRAM and P3 editing, on which J.S. is listed as inventor. J.S. is on the scientific advisory board, a consultant, and/or a co-founder of Adaptive Biotechnologies, Cajal Neuroscience, Camp4 Therapeutics, Guardant Health, Maze Therapeutics, Pacific Biosciences, Phase Genomics, Prime Medicine, Scale Biosciences, and Sixth Street Capital.
Figures


References
-
- Alcolea MP, Jones PH. Tracking cells in their native habitat: lineage tracing in epithelial neoplasia. Nat Rev Cancer. 2013. Mar;13(3):161–71. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

