Identification and validation of a costimulatory molecule-related signature to predict the prognosis for uveal melanoma patients
- PMID: 38644411
- PMCID: PMC11033288
- DOI: 10.1038/s41598-024-59827-5
Identification and validation of a costimulatory molecule-related signature to predict the prognosis for uveal melanoma patients
Abstract
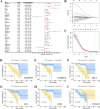
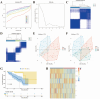
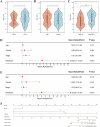
Uveal melanoma (UVM) is the most common primary tumor in adult human eyes. Costimulatory molecules (CMs) are important in maintaining T cell biological functions and regulating immune responses. To investigate the role of CMs in UVM and exploit prognostic signature by bioinformatics analysis. This study aimed to identify and validate a CMs associated signature and investigate its role in the progression and prognosis of UVM. The expression profile data of training cohort and validation cohort were downloaded from The Cancer Genome Atlas (TCGA) dataset and the Gene Expression Omnibus (GEO) dataset. 60 CM genes were identified, and 34 genes were associated with prognosis by univariate Cox regression. A prognostic signature was established with six CM genes. Further, high- and low-risk groups were divided by the median, and Kaplan-Meier (K-M) curves indicated that high-risk patients presented a poorer prognosis. We analyzed the correlation of gender, age, stage, and risk score on prognosis by univariate and multivariate regression analysis. We found that risk score was the only risk factor for prognosis. Through the integration of the tumor immune microenvironment (TIME), it was found that the high-risk group presented more immune cell infiltration and expression of immune checkpoints and obtained higher immune scores. Enrichment analysis of the biological functions of the two groups revealed that the differential parts were mainly related to cell-cell adhesion, regulation of T-cell activation, and cytokine-cytokine receptor interaction. No differences in tumor mutation burden (TMB) were found between the two groups. GNA11 and BAP1 have higher mutation frequencies in high-risk patients. Finally, based on the Genomics of Drug Sensitivity in Cancer 2 (GDSC2) dataset, drug sensitivity analysis found that high-risk patients may be potential beneficiaries of the treatment of crizotinib or temozolomide. Taken together, our CM-related prognostic signature is a reliable biomarker that may provide ideas for future treatments for the disease.
Keywords: Biomarker; Costimulatory molecular; Prognosis signature; Tumor immune microenvironment; Uveal melanoma.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






Similar articles
-
A Comprehensive Prognostic and Immunological Analysis of a Six-Gene Signature Associated With Glycolysis and Immune Response in Uveal Melanoma.Front Immunol. 2021 Sep 22;12:738068. doi: 10.3389/fimmu.2021.738068. eCollection 2021. Front Immunol. 2021. PMID: 34630418 Free PMC article.
-
A Golgi apparatus-related signature for predicting prognosis and evaluating the tumor immune microenvironment of uveal melanoma.Medicine (Baltimore). 2025 Jun 20;104(25):e42909. doi: 10.1097/MD.0000000000042909. Medicine (Baltimore). 2025. PMID: 40550052 Free PMC article.
-
Development and Validation of an Extracellular Matrix Gene Expression Signature for Prognostic Prediction in Patients with Uveal Melanoma.Int J Mol Sci. 2025 May 1;26(9):4317. doi: 10.3390/ijms26094317. Int J Mol Sci. 2025. PMID: 40362553 Free PMC article.
-
Single cell RNA-sequencing in uveal melanoma: advances in heterogeneity, tumor microenvironment and immunotherapy.Front Immunol. 2024 Jun 20;15:1427348. doi: 10.3389/fimmu.2024.1427348. eCollection 2024. Front Immunol. 2024. PMID: 38966635 Free PMC article. Review.
-
A Novel Pancreatic Cancer Hypoxia Status Related Gene Signature for Prognosis and Therapeutic Responses.Mol Biotechnol. 2024 Jul;66(7):1684-1703. doi: 10.1007/s12033-023-00807-x. Epub 2023 Jul 5. Mol Biotechnol. 2024. PMID: 37405638 Review.
Cited by
-
Development of a prognostic nomogram for ocular melanoma: a SEER population-based study (2000-2021).Front Med (Lausanne). 2025 Jan 29;12:1494925. doi: 10.3389/fmed.2025.1494925. eCollection 2025. Front Med (Lausanne). 2025. PMID: 39944484 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

