Dynamic changes in the plastid and mitochondrial genomes of the angiosperm Corydalis pauciovulata (Papaveraceae)
- PMID: 38644497
- PMCID: PMC11034061
- DOI: 10.1186/s12870-024-05025-4
Dynamic changes in the plastid and mitochondrial genomes of the angiosperm Corydalis pauciovulata (Papaveraceae)
Abstract
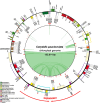
Background: Corydalis DC., the largest genus in the family Papaveraceae, comprises > 465 species. Complete plastid genomes (plastomes) of Corydalis show evolutionary changes, including syntenic arrangements, gene losses and duplications, and IR boundary shifts. However, little is known about the evolution of the mitochondrial genome (mitogenome) in Corydalis. Both the organelle genomes and transcriptomes are needed to better understand the relationships between the patterns of evolution in mitochondrial and plastid genomes.
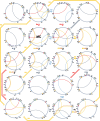
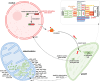
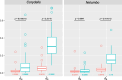
Results: We obtained complete plastid and mitochondrial genomes from Corydalis pauciovulata using a hybrid assembly of Illumina and Oxford Nanopore Technologies reads to assess the evolutionary parallels between the organelle genomes. The mitogenome and plastome of C. pauciovulata had sizes of 675,483 bp and 185,814 bp, respectively. Three ancestral gene clusters were missing from the mitogenome, and expanded IR (46,060 bp) and miniaturized SSC (202 bp) regions were identified in the plastome. The mitogenome and plastome of C. pauciovulata contained 41 and 67 protein-coding genes, respectively; the loss of genes was a plastid-specific event. We also generated a draft genome and transcriptome for C. pauciovulata. A combination of genomic and transcriptomic data supported the functional replacement of acetyl-CoA carboxylase subunit β (accD) by intracellular transfer to the nucleus in C. pauciovulata. In contrast, our analyses suggested a concurrent loss of the NADH-plastoquinone oxidoreductase (ndh) complex in both the nuclear and plastid genomes. Finally, we performed genomic and transcriptomic analyses to characterize DNA replication, recombination, and repair (DNA-RRR) genes in C. pauciovulata as well as the transcriptomes of Liriodendron tulipifera and Nelumbo nuicifera. We obtained 25 DNA-RRR genes and identified their structure in C. pauciovulata. Pairwise comparisons of nonsynonymous (dN) and synonymous (dS) substitution rates revealed that several DNA-RRR genes in C. pauciovulata have higher dN and dS values than those in N. nuicifera.
Conclusions: The C. pauciovulata genomic data generated here provide a valuable resource for understanding the evolution of Corydalis organelle genomes. The first mitogenome of Papaveraceae provides an example that can be explored by other researchers sequencing the mitogenomes of related plants. Our results also provide fundamental information about DNA-RRR genes in Corydalis and their related rate variation, which elucidates the relationships between DNA-RRR genes and organelle genome stability.
Keywords: Concomitant loss; DNA-RRR; Genome rearrangement; NDH complex; Organelle genomes.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Contrasting Patterns of Nucleotide Substitution Rates Provide Insight into Dynamic Evolution of Plastid and Mitochondrial Genomes of Geranium.Genome Biol Evol. 2017 Jun 1;9(6):1766-1780. doi: 10.1093/gbe/evx124. Genome Biol Evol. 2017. PMID: 28854633 Free PMC article.
-
Coevolution between Nuclear-Encoded DNA Replication, Recombination, and Repair Genes and Plastid Genome Complexity.Genome Biol Evol. 2016 Feb 17;8(3):622-34. doi: 10.1093/gbe/evw033. Genome Biol Evol. 2016. PMID: 26893456 Free PMC article.
-
Rate accelerations in plastid and mitochondrial genomes of Cyperaceae occur in the same clades.Mol Phylogenet Evol. 2023 May;182:107760. doi: 10.1016/j.ympev.2023.107760. Epub 2023 Mar 13. Mol Phylogenet Evol. 2023. PMID: 36921696
-
Plastomes on the edge: the evolutionary breakdown of mycoheterotroph plastid genomes.New Phytol. 2017 Apr;214(1):48-55. doi: 10.1111/nph.14398. Epub 2017 Jan 9. New Phytol. 2017. PMID: 28067952 Review.
-
Invited Review Beyond parasitic convergence: unravelling the evolution of the organellar genomes in holoparasites.Ann Bot. 2023 Nov 30;132(5):909-928. doi: 10.1093/aob/mcad108. Ann Bot. 2023. PMID: 37503831 Free PMC article. Review.
Cited by
-
Comparative analysis of mitochondrial and chloroplast genomes of Dracaena cambodiana from contrasting island habitats.Front Plant Sci. 2025 Jun 25;16:1620721. doi: 10.3389/fpls.2025.1620721. eCollection 2025. Front Plant Sci. 2025. PMID: 40636010 Free PMC article.
-
Deciphering the complex organelle genomes of two Rhododendron species and insights into adaptive evolution patterns in high-altitude.BMC Plant Biol. 2024 Nov 8;24(1):1054. doi: 10.1186/s12870-024-05761-7. BMC Plant Biol. 2024. PMID: 39511517 Free PMC article.
-
Elucidating the evolutionary dynamics of parasitism in Cuscuta: in-depth phylogenetic reconstruction and extensive plastomes reduction.BMC Genomics. 2025 Feb 12;26(1):137. doi: 10.1186/s12864-025-11324-3. BMC Genomics. 2025. PMID: 39939920 Free PMC article.
-
Beyond conservation: the landscape of chloroplast genome rearrangements in angiosperms.New Phytol. 2025 Sep;247(6):2571-2580. doi: 10.1111/nph.70364. Epub 2025 Jul 4. New Phytol. 2025. PMID: 40613318 Free PMC article. Review.
-
Comparative analysis of mitochondrial genomes in lycoperdaceae fungi reveals intron dynamics and phylogenetic relationships.BMC Genomics. 2025 Aug 11;26(1):742. doi: 10.1186/s12864-025-11911-4. BMC Genomics. 2025. PMID: 40790547 Free PMC article.
References
-
- Mower JP, Sloan DB, Alverson AJ. Plant mitochondrial genome diversity: the genomics revolution. In: Wendel JH , editor. Plant genome diversity volume 1: plant genomes, their residents, and their evolutionary dynamics. New York: Springer; 2012. p. 123–144.
-
- Ruhlman TA, Jansen RK. Plastid genomes of flowering plants: essential principles. In: Maliga P, editor. Chloroplast biotechnology: methods and protocols. New York: Springer, US; 2021. pp. 3–47. - PubMed
MeSH terms
Grants and funding
- 2017R1A6A3A11034431/the National Research Foundation of Korea (NRF) funded by the Ministry of Education
- 2017R1A6A3A11034431/the National Research Foundation of Korea (NRF) funded by the Ministry of Education
- 2017R1A6A3A11034431/the National Research Foundation of Korea (NRF) funded by the Ministry of Education
LinkOut - more resources
Full Text Sources

